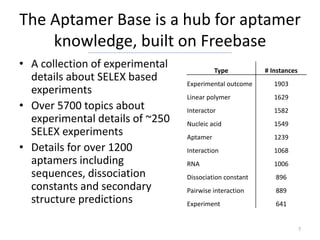
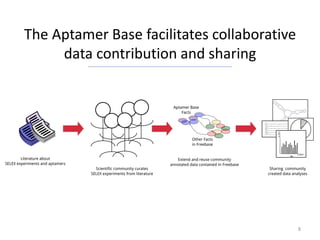
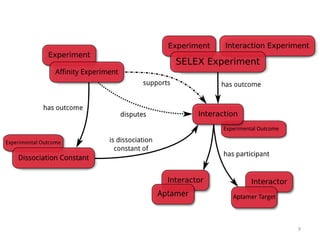
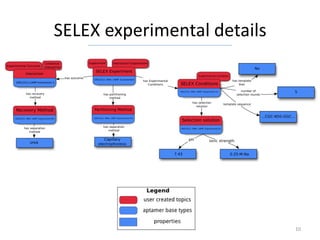
The Aptamer Base is a collaborative knowledgebase designed to document aptamers and SELEX experiments, offering a comprehensive collection of experimental data and interaction details. It addresses the limitations of existing aptamer databases by allowing user contributions and facilitating advanced querying through Metaweb Query Language. Future plans include enhancing data entry interfaces and integrating additional research publications.

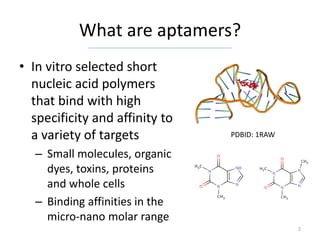


![MQL allows powerful querying over
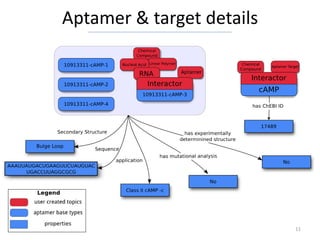
the Aptamer Base Find all affinity experiments that dispute interactions that have as participant
How many SELEX experiments are described “Sulforhodamine b”
in the Aptamer Base? [{
[{ "mid": null,
"type": "/base/aptamer/selex_experiment", "type": "/base/aptamer/affinity_experiment",
"disputes": [{
"return": "count" "type": "/base/aptamer/interaction",
}] "has_participant": [{
Results: http://tinyurl.com/d3ox66m "type": "/base/aptamer/molecule",
"name": "Sulforhodamine b"
Provide a list of all interactions that have as
}]
participants RNA aptamers
}]
[{ }]
Results: http://tinyurl.com/bmcyfdw
"id": null,
"type": "/base/aptamer/interaction", Find all dissociation constants that have a value that is less than 1E-6 for
interactions involving either L-Arginine or Thrombin
"has_participant": [{ [{
"mid": null,
"id": null, "type": "/base/aptamer/dissociation_constant",
"has_value<": 0.000001,
"a:type": "/base/aptamer/aptamer", "/base/aptamer/dissociation_constant/is_dissociation_constant_of": [{
"b:type": "/base/aptamer/rna" "/base/aptamer/interaction/has_participant": [{
"mid": null,
}] "type": "/base/aptamer/aptamer_target",
"name|=": [
}] "L-Arginine",
"Thrombin"
Results: http://tinyurl.com/cckdax7 ]
}]
}] Results: http://tinyurl.com/c8ystcg
}]
13](https://image.slidesharecdn.com/cruztoledo-aptamerbasebc2012-120428024723-phpapp02/85/Jose-Cruz-Toledo-Aptamer-basebc2012-13-320.jpg)
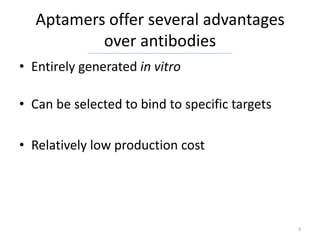
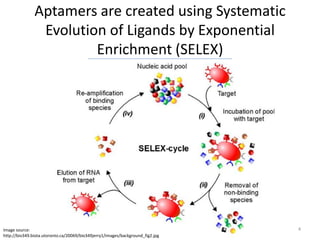
![The Aptamer Base is not just another
Aptamer Database
• The Aptamer Base is the only collaborative
aptamer resource on the web
• Users are not limited to an HTML form for
accessing data
– TSV :
http://download.freebase.com/datadumps/latest/browse/base/aptamer/
– RDF:
http://rdf.freebase.com/rdf/[TOPIC_ID]
– Powerful query language:
http://wiki.freebase.com/wiki/MQL
• The aptamer Base extends over ~2700 pre-
existing topics on Freebase
14](https://image.slidesharecdn.com/cruztoledo-aptamerbasebc2012-120428024723-phpapp02/85/Jose-Cruz-Toledo-Aptamer-basebc2012-14-320.jpg)