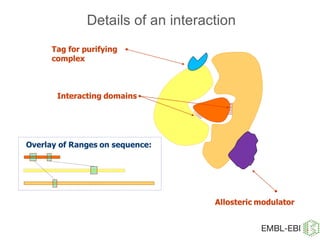
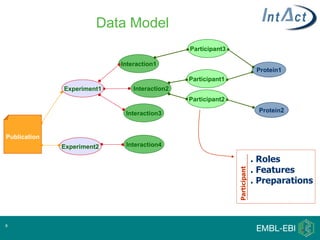
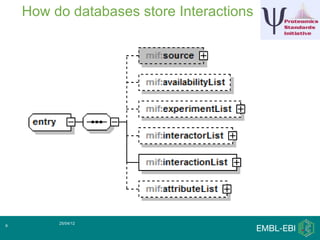
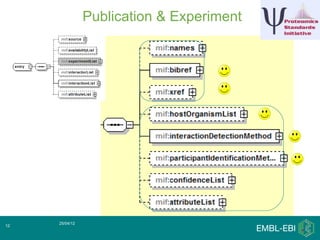
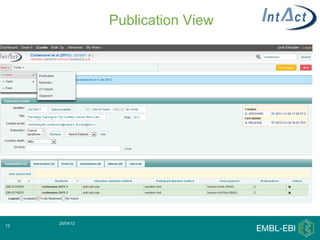
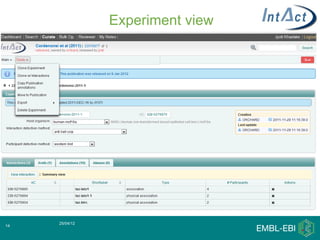
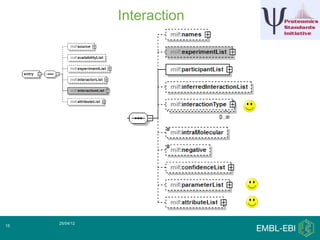
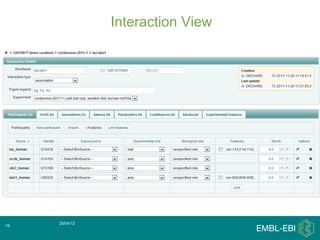
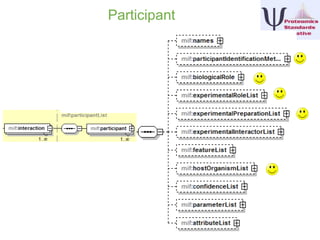
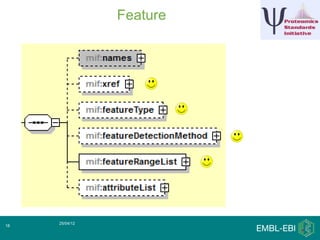
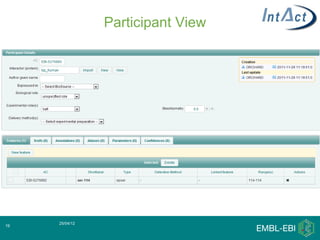
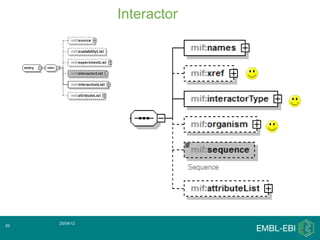
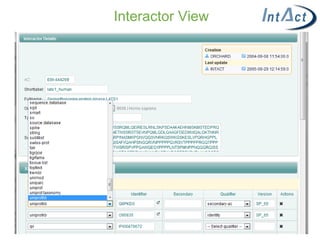
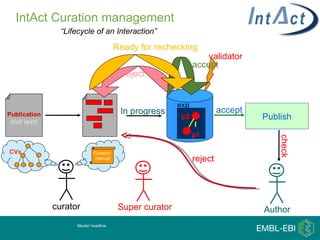
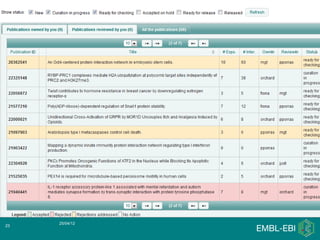
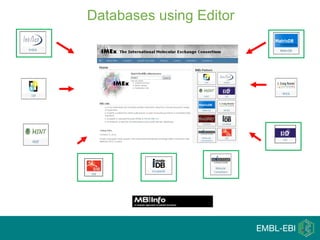
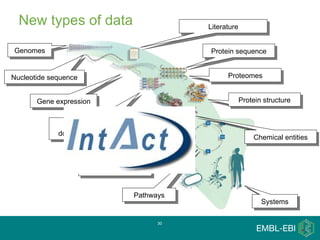
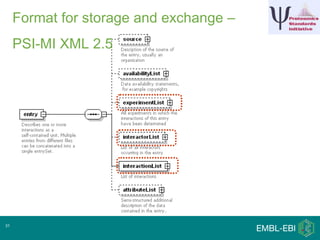
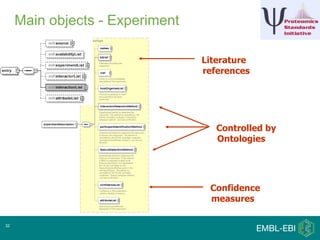
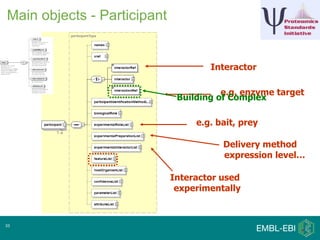
IntAct is a database that curates molecular interaction data manually from literature. It uses the IntAct editor, which is based on the PSI-MI XML schema, to capture interaction details such as participating proteins, experimental conditions, and interaction types. The editor allows curators to manage the curation process and stores the data in a standardized format that can be shared with other databases.