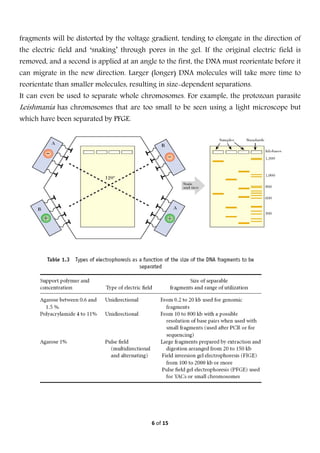
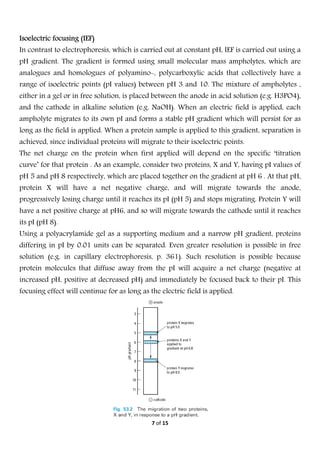
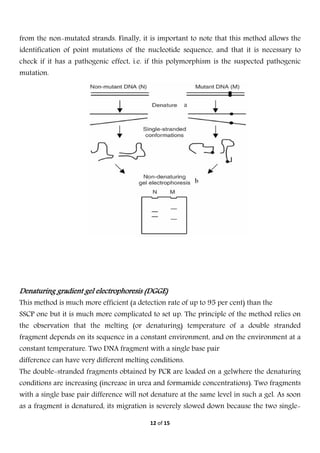
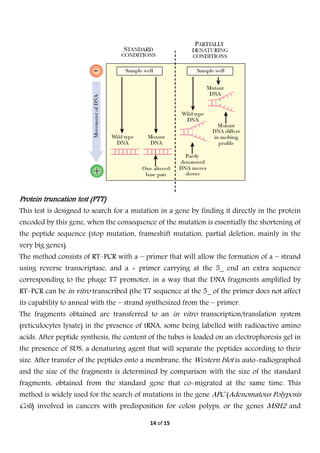
Electrophoresis is a technique used to separate molecules based on their charge and size. It works by applying an electric field to move charged molecules through a medium like agarose gel or polyacrylamide gel at different rates. Key factors that determine a molecule's movement include its net charge, size, shape, and the strength of the electric field. Variations of electrophoresis, like isoelectric focusing and two-dimensional electrophoresis, allow for highly precise separations of proteins and other biomolecules. Emerging techniques like pulsed field gel electrophoresis and capillary electrophoresis provide even higher resolution.