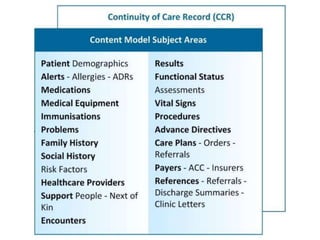
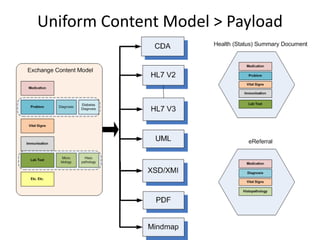
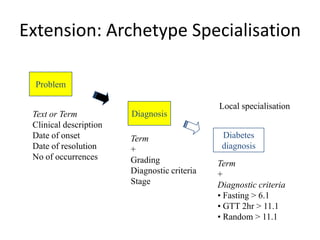
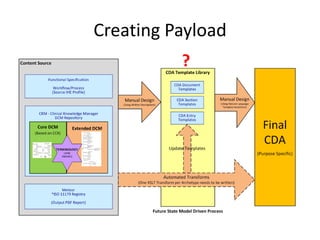
This document provides a first draft of a content model for health information exchange (HIE). It describes reusable clinical information bits called archetypes organized following the CCR standard. International and local archetypes as well as extensions can be included. When creating HIE payloads, only relevant archetypes would be included. The document discusses developing content through grassroots and governance processes and demonstrates example HIE models and capabilities.