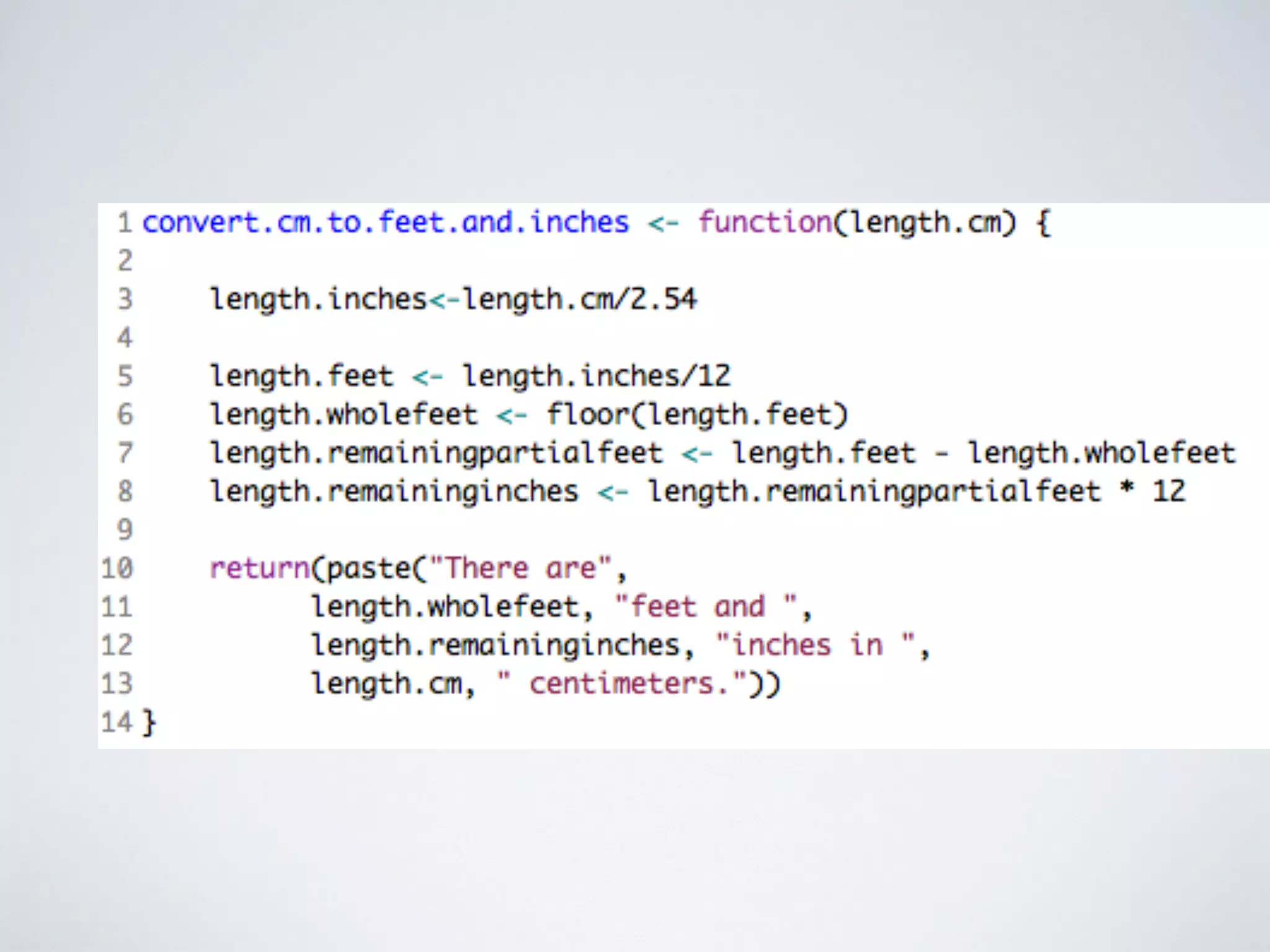
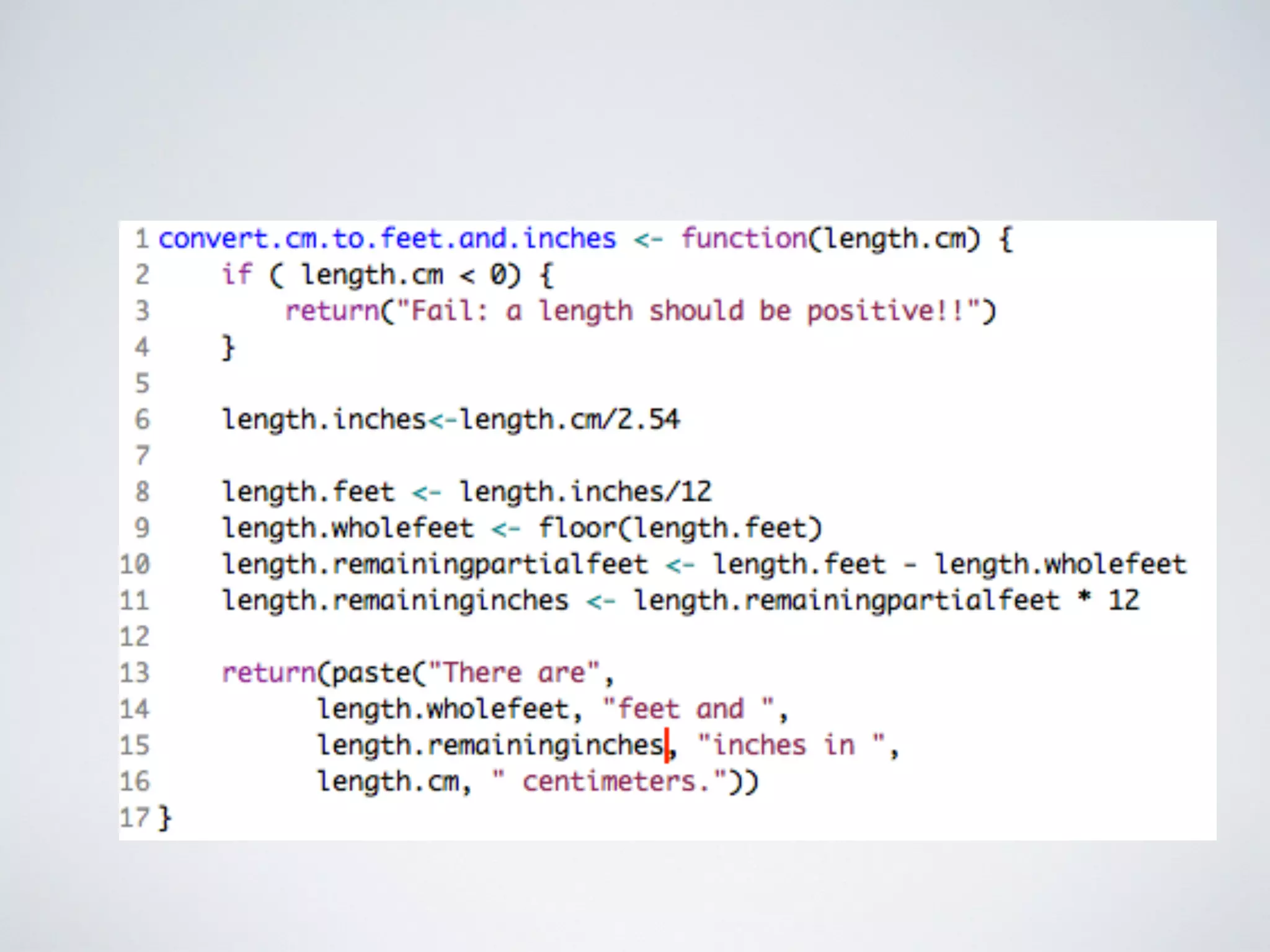
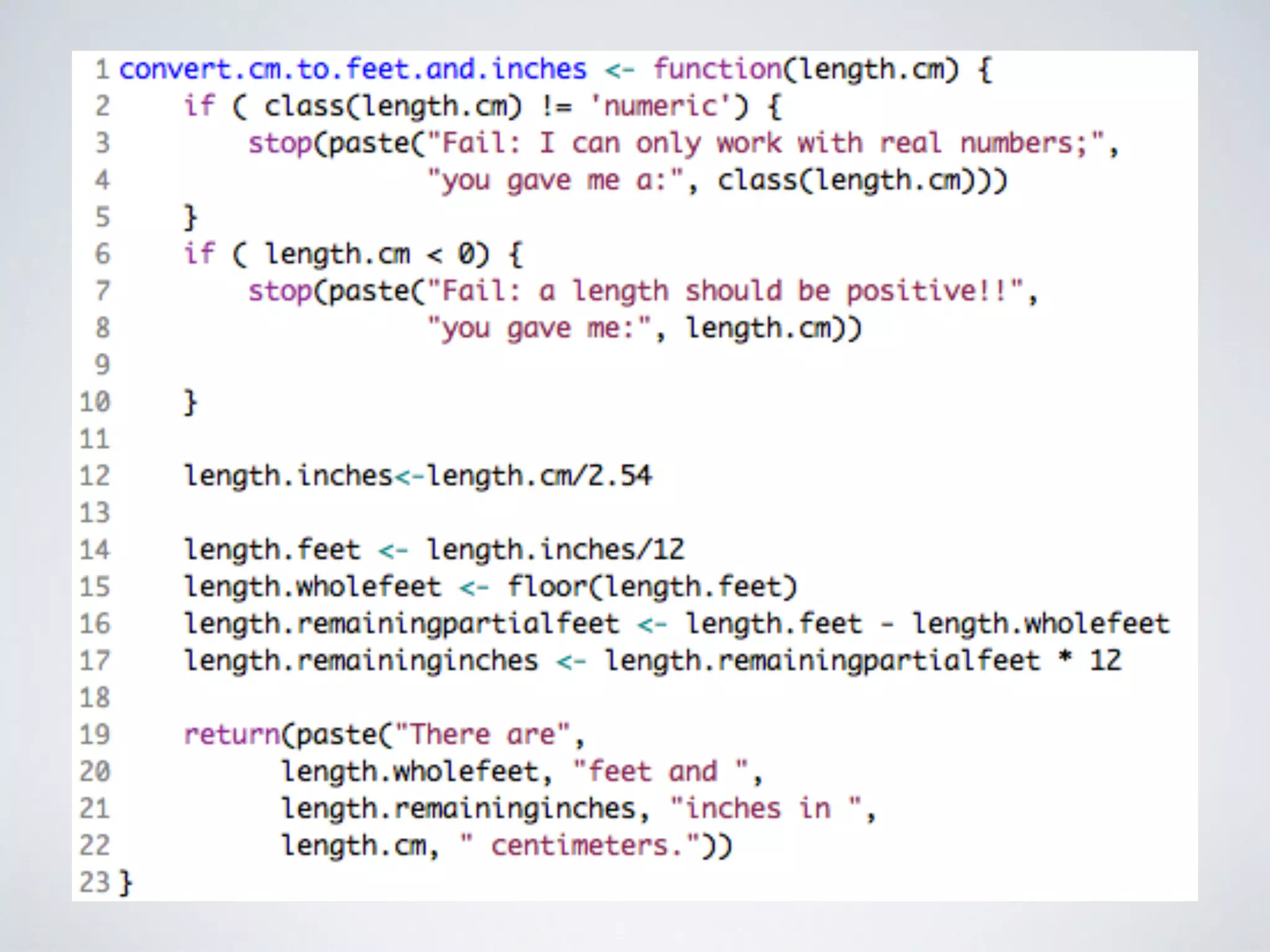
This document contains information about programming in R, including practical examples. It discusses accessing and subsetting data, using regular expressions for text search, creating functions, and using loops. Examples are provided to demonstrate creating vectors, accessing subsets of vectors, using regular expressions to find patterns in text, creating functions to convert between units or estimate values, and using for loops to repeat operations over multiple elements. The document suggests R is useful for working with big data in biology and other fields due to its ability to automate tasks, integrate with other tools, and handle large datasets through programming.

























![• creating a vector
> my_vector <- c(5, 6, 7, 8, 9, 10, 11)
> my_vector <- 5:11
> my_vector <- seq(from=5, to=11, by=1)
> my_vector
[1] 5 6 7 8 9 10 11
> length(my_vector)
[1] 7
> (10 > 30)
[1] FALSE
> my_vector > 8
[1] FALSE FALSE FALSE FALSE TRUE TRUE TRUE
> my_vector[my_vector > 8]
9 10 11
> other_vector <- my_vector[my_vector > 8]
> other_vector
9 10 11
> other_vector + 3
• give me a vector containing numbers from 5 to 11 (3 variants)](https://image.slidesharecdn.com/2015-11-17-programminginr-151117212739-lva1-app6891/75/2015-11-17-programming-inr-key-36-2048.jpg)
![• accessing a subset
• of a vector
> big_vector <- 150:100
> big_vector
[1] 150 149 148 147 146 145 144 143 142 141 140 139 138 137 136 135 13
[20] 131 130 129 128 127 126 125 124 123 122 121 120 119 118 117 116 11
[39] 112 111 110 109 108 107 106 105 104 103 102 101 100
> big_vector[5]
146
> mysubset <- big_vector[my_vector]
> mysubset
[1] 146 145 144 143 142 141 140
> big_vector > 130
[1] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
[13] TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE
[25] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[37] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[49] FALSE FALSE FALSE
> subset(x = big_vector, subset = big_vector > 140)
[1] 150 149 148 147 146 145 144 143 142 141
> big_vector[big_vector >= 140]
[1] 150 149 148 147 146 145 144 143 142 141 140
> my_vector
[1] 5 6 7 8 9 10 11](https://image.slidesharecdn.com/2015-11-17-programminginr-151117212739-lva1-app6891/75/2015-11-17-programming-inr-key-37-2048.jpg)




![Regular expressions (regex):
Text search on steroids.
Regular expression Finds
David David
Dav(e|(id)) David, Dave
Dav(e|(id)|(ide)|o) David, Dave, Davide, Davo
At{1,2}enborough
Attenborough,
Atenborough
Atte[nm]borough
Attenborough,
Attemborough
At{1,2}[ei][nm]bo{0,1}ro((ugh)|w){0,1}
Atimbro,
attenbrough,
ateinborow
Easy counting, replacing all with “Sir David Attenborough”
Yes: ”HATSOMIKTIP"
yes: ”HAVSONYYIKTIP"
not: ”HAVSQMIKTIP"](https://image.slidesharecdn.com/2015-11-17-programminginr-151117212739-lva1-app6891/75/2015-11-17-programming-inr-key-42-2048.jpg)
![Regex special symbols
Regular expression Finds Example
[aeiou] any single vowel “e”
[aeiou]*
between 0 and infinity
vowels vowels, e.g.’
“eeooouuu"
[aeoiu]{1,3} between 1 and 3 vowels “oui”
a|i one of the 2 characters “"
((win)|(fail))
one of the two
words in ()
fail
Yes: ”HATSOMIKTIP"
yes: ”HAVSONYYIKTIP"
not: ”HAVSQMIKTIP"](https://image.slidesharecdn.com/2015-11-17-programminginr-151117212739-lva1-app6891/75/2015-11-17-programming-inr-key-43-2048.jpg)
![More Regex Special symbols
• Google “Regular expression cheat sheet”
• ?regexp
Synonymous with
[:digit:] [0-9]
[A-z] [A-z], ie [A-Za-z]
s whitespace
. any single character
.+ one to many of anything
b* between 0 and infinity letter ‘b’
[^abc] any character other than a, b or c.
( (
[:punct:]
any of these: ! " # $ % & ' ( ) * + , - . /
: ; < = > ? @ [ ] ^ _ ` { |](https://image.slidesharecdn.com/2015-11-17-programminginr-151117212739-lva1-app6891/75/2015-11-17-programming-inr-key-44-2048.jpg)










![Functions
• R has many. e.g.: plot(), t.test()
• Making your own:
tree_age_estimate <- function(diameter, species) {
growth_rate <- growth_rates[ species ]
age_estimate <- diameter / growth_rate
return(age_estimate)
}
> tree_age_estimate(25, “White Oak”)
+ 66
> tree_age_estimate(60, “Carya ovata”)
+ 190](https://image.slidesharecdn.com/2015-11-17-programminginr-151117212739-lva1-app6891/75/2015-11-17-programming-inr-key-55-2048.jpg)

![“for”
Loop
> possible_colours <- c('blue', 'cyan', 'sky-blue', 'navy blue',
'steel blue', 'royal blue', 'slate blue', 'light blue', 'dark
blue', 'prussian blue', 'indigo', 'baby blue', 'electric blue')
> possible_colours
[1] "blue" "cyan" "sky-blue" "navy blue"
[5] "steel blue" "royal blue" "slate blue" "light blue"
[9] "dark blue" "prussian blue" "indigo" "baby blue"
[13] "electric blue"
> for (colour in possible_colours) {
+ print(paste("The sky is oh so, so", colour))
+ }
[1] "The sky is so, oh so blue"
[1] "The sky is so, oh so cyan"
[1] "The sky is so, oh so sky-blue"
[1] "The sky is so, oh so navy blue"
[1] "The sky is so, oh so steel blue"
[1] "The sky is so, oh so royal blue"
[1] "The sky is so, oh so slate blue"
[1] "The sky is so, oh so light blue"
[1] "The sky is so, oh so dark blue"
[1] "The sky is so, oh so prussian blue"
[1] "The sky is so, oh so indigo"
[1] "The sky is so, oh so baby blue"](https://image.slidesharecdn.com/2015-11-17-programminginr-151117212739-lva1-app6891/75/2015-11-17-programming-inr-key-58-2048.jpg)

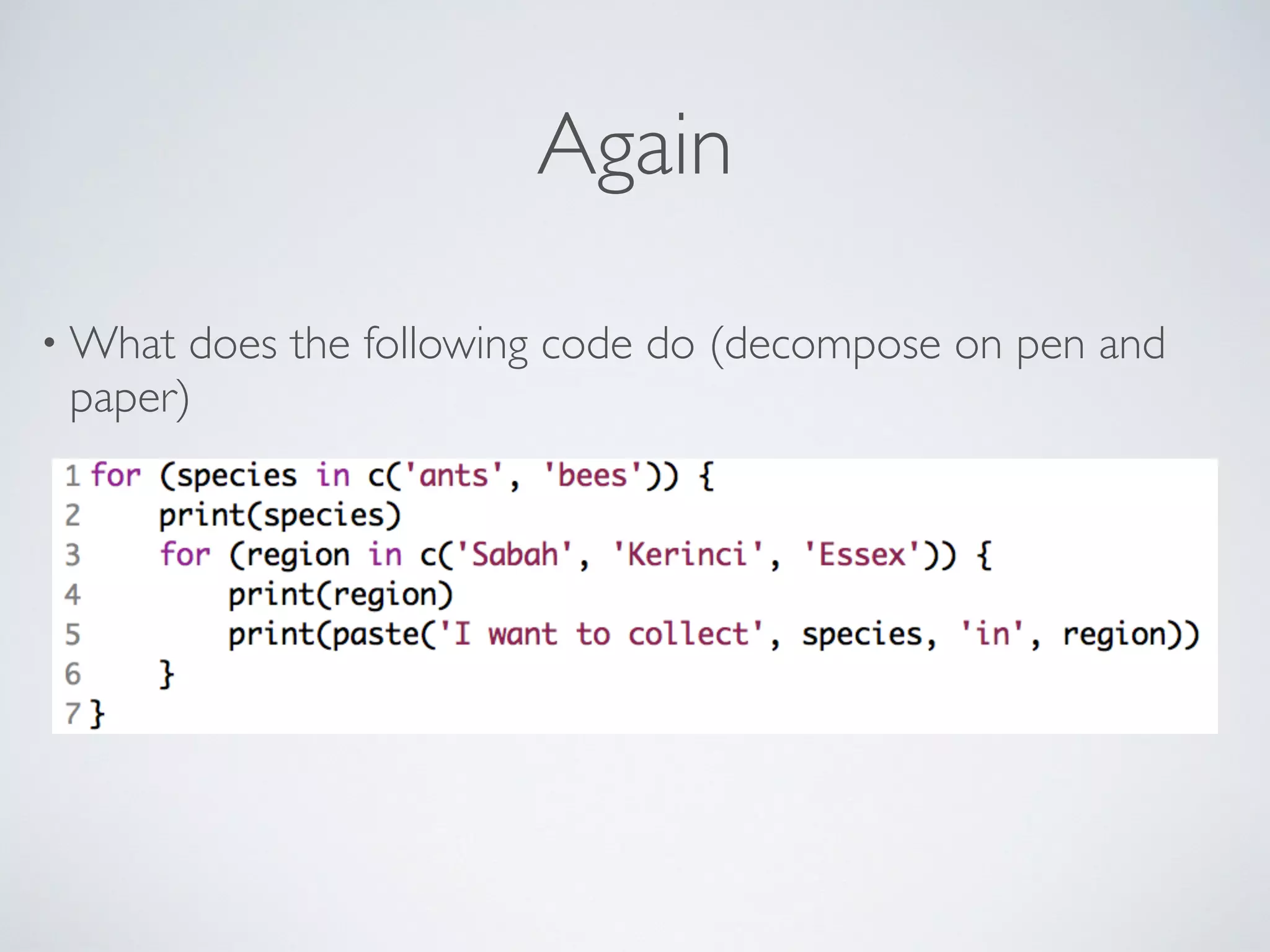
![for (letter in LETTERS) {
begins_with <- paste("^", letter, sep="")
matches <- grep(pattern = begins_with,
x = ants$genus)
print(paste(length(matches), "begin with", letter))
}
> LETTERS
[1] "A" "B" "C" "D" "E" "F" "G" "H" "I" "J" "K" "L" "M" "N" "O" "P" "Q" "R" "S"
[20] "T" "U" "V" "W" "X" "Y" "Z"
> ants <- read.table("https://goo.gl/3Ek1dL")
> colnames(ants) <- c("genus", “species")
> head(ants)
genus species
1 Anergates atratulus
2 Camponotus sp.
3 Crematogaster scutellaris
4 Formica aquilonia
5 Formica cunicularia
6 Formica exsecta
What does this loop do?](https://image.slidesharecdn.com/2015-11-17-programminginr-151117212739-lva1-app6891/75/2015-11-17-programming-inr-key-61-2048.jpg)