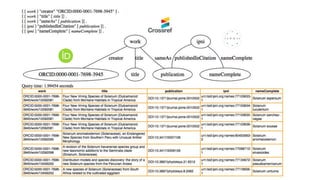
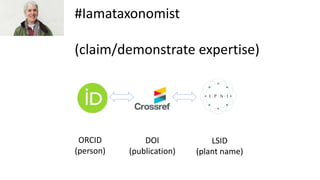
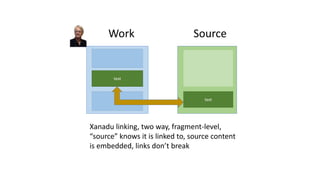
The document discusses obstacles to and progress in building knowledge graphs. It outlines technical obstacles like needing globally unique identifiers and agreed-upon vocabularies. Social obstacles include economic issues. Identifiers are key to connecting knowledge graphs. Metrics are needed to measure graph connectivity rather than just growth. Network effects are important to make graphs truly useful. Wikidata and Google's knowledge graph are examples of existing large knowledge graphs.



















![Hexastore
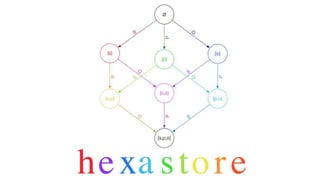
• A triple is [s, p, o]
• Find all statements [s, ?, ?] is simple array lookup (all elements with key “s”)
• Find all statements [?, ?, o] is slow (scan all triples)…
• …unless we add array of [o, s, p] triples, then simple array lookup (all elements with
key “o”)
• Six variations cover all queries: [s,p,o], [s,o,p], [p, s, o], [p, o, s], [o, s, p], [o, p, s]
(hence “hexastore”)
• In-memory graph database in Javascript (think offline apps)
http://crubier.github.io/Hexastore/](https://image.slidesharecdn.com/rdmp-keynote-171002200740/85/Towards-a-biodiversity-knowledge-graph-24-320.jpg)








![Embedded markup (bad)…
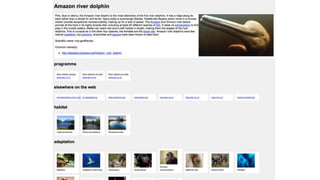
Crocidura absconditus, new species
<i>Crocidura absconditus</i>, new species
0 20
{ [0,20], “italics” }
…versus annotation (good)
(think NLM JATS XML markup
versus Substance JSON used
by Lens viewer
https://lens.elifesciences.org/
about/)
Crocidura absconditus, new species](https://image.slidesharecdn.com/rdmp-keynote-171002200740/85/Towards-a-biodiversity-knowledge-graph-36-320.jpg)