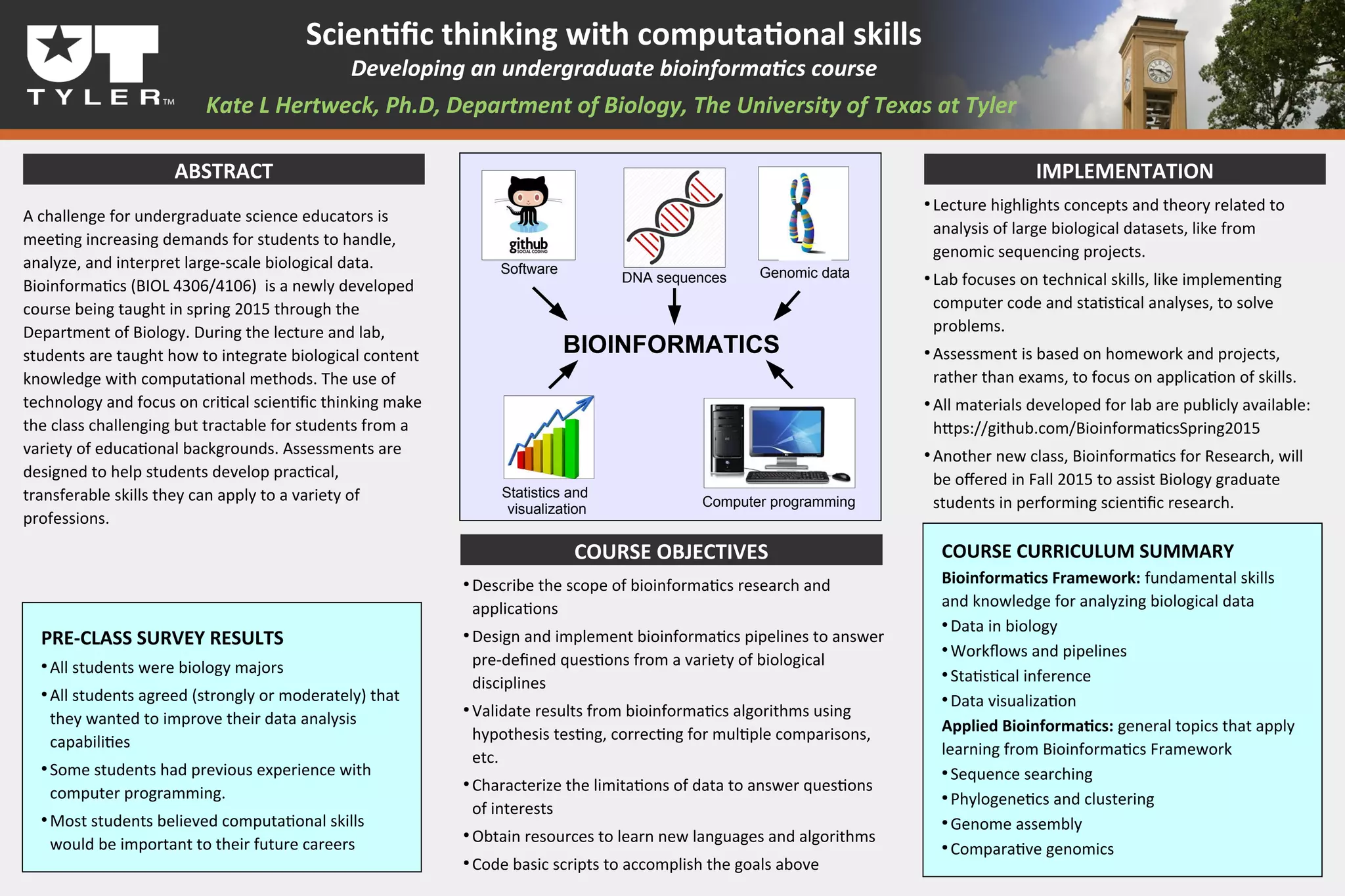
The document describes a newly developed undergraduate bioinformatics course at the University of Texas at Tyler. The course teaches students how to integrate biological knowledge with computational methods to handle and analyze large-scale biological data. Students learn technical skills like computer coding and statistical analysis in labs, and are assessed through projects rather than exams to focus on applying their skills. The course aims to help students develop practical and transferable competencies for various professions.