Drug discovery algorithm identifies REL1 inhibitors
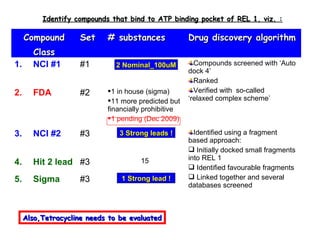
- 1. CompoundCompound ClassClass SetSet # substances# substances Drug discovery algorithmDrug discovery algorithm 1. NCI #1 #1 16 Compounds screened with ‘Auto dock 4’ Ranked Verified with so-called ‘relaxed complex scheme’ 2. FDA #2 1 in house (sigma) 11 more predicted but financially prohibitive 1 pending (Dec 2009) 3. NCI #2 #3 8 Identified using a fragment based approach: Initially docked small fragments into REL 1 Identified favourable fragments Linked together and several databases screened 4. Hit 2 lead #3 15 5. Sigma #3 2 Identify compounds that bind to ATP binding pocket of REL 1, viz. :Identify compounds that bind to ATP binding pocket of REL 1, viz. : 1 Strong lead !1 Strong lead ! 3 Strong leads !3 Strong leads ! 2 Nominal_100uM2 Nominal_100uM Also,Tetracycline needs to be evaluatedAlso,Tetracycline needs to be evaluated
- 2. CompoundCompound TypeType CompoundCompound 1010µµMM 100100µMµM TitratedTitrated N=N= ICIC5050 RangeRange CommentsComments NCI BatchNCI Batch #2#2 # 45609 ! 95 % (78%) (88%) 33 3-1uM Discrepancy between assays above predicted IC50 range Repeat ? Discrepancy between assays Repeated ! (#0036) # 162535 !!# 162535 !! 85_90 % (85%) (~98%)* (90 %) (>99.9%)* >3>3 (+2)(+2) ~1uM (N=1) ~3-1uM (N=3) # 1698 ! 80 % (50%) (90%) 33 (+2)(+2) 10uM- 3uM (N=2) 3-1uM (N=1) NCI BatchNCI Batch #1#1 # 117079 30_80% >99% (~80 %) (~50-70%) >3>3 100uM- 30uM (N=4) 30uM (N=1) Discrepancy between assays above IC50 range Furthermore, IC50’s differ between assays Repeat ? # 125908 20_70% ~95 % (~50-70%) >3>3 100uM- 30uM (N=3) ~30uM (N=1) # 45201 NA ~70% Yet to do !!Yet to do !! Sigma rareSigma rare LibraryLibrary MordantMordant Black !!Black !! (~98%)* (~99.8%)* >3>3 ~1uM Completed Values derive from titrations’ Identified using fragment based algorithm Identified using Auto Dock 4 relaxed complex scheme Assay #0036 is an attempt to clarify IC50’s for # 1698 & #162535 Regarding # 45609 IC50’s concur but values differ above this threshold Regarding #117079 & #125908 some assays have different IC50’s Furthermore, those with similar values differ above IC50 titrant * Values derived from 0.25ul undiluted prep #1
- 3. 34B MBMB #162535#162535 # 45609# 45609 # 1698# 1698 1uM1uM >/=1uM>/=1uM 3-1uM3-1uM >3uM>3uM
- 4. % (Putative) Inhibition of REL 1 Ligation (Adenylation) by Mordant Black (solute) relative to DMSO (solvent)_# Mordant Black Molarity Titration 100uM 100uM 30uM 30uM 10uM 10uM DMSO DMSO 3uM 3uM 1uM 1uM 300nM 300nM 100nM 100nM 30nM 30nM 0 20 40 60 80 100 120 140 Prep #3_050203 Prep #1_080802 Mordant Black Molarity Titration: 100uM-30nM SpecificSignal/DMSO_% 3.4%3.4% 10%10% 23.7%23.7% 52%52% 81%81% <0.5%<0.5% 5%5% 26.5%26.5% 55.8%55.8% 75%75% 1ul Enzyme1ul Enzyme 0.25ul Enzyme0.25ul Enzyme Although both REL 1 BAC preps differ with regard to intrinsic activity both yield putative IC50’s approx to 1uM This concurs with prior assays100uM100uM 30nM30nM 50S50S-1-1 100S100S-1-1
- 5. % (Putative) Inhibition of Ligation (Adenylation) by Mordant Black (solute) relative to DMSO (solvent)_# Mordant Black Molarity Titration 30uM 30uM 10uM 10uM 10uM 100uM 100uM 100uM 30uM 0 20 40 60 80 100 120 140 Prep #3_050203 Prep #1_080802 Prep #1_080802 Mordant Black Molarity Titration: 100uM-30nM SpecificSignal/DMSO_% # 0029A # 0033A 28,500,000 152,500,000 203,500,000 52% 56% 54% 1ul 50% REL 1 0.25ul 50% REL 1 1ul undiluted REL 1 17 HR17 HR 1 HR ! For assays depicted (N=3) IC50 consistently approximates to ~1uM Undiluted prep #1 displays massively greater signal than either glycerol diluted #1 or #3
- 6. % (Putative) Inhibition of Ligation (Adenylation) by Mordant Black (solute) relative to DMSO (solvent)_# Mordant Black Molarity Titration DMSO DMSO 100uM 100uM 30uM 30uM 10uM 10uM 3uM 3uM 1uM 1uM 300nM 300nM 100nM 100nM 30nM 30nM 0 20 40 60 80 100 120 140 Prep #1_080802 Prep #3_050203 Molarity Titration: 100uM-30nM SpecificSignal/DMSO_% 24+/-5.6% 59+/-5.2% 24% 52% IC50 approximates to 1uM for N=5 Combining data from assays #0029_33_34 culminates in predicted IC50 of ~1uM (N=4) In terms of IC50 alone this is consistent with data from REL 1 prep #3 also derived from #0029
- 7. % (Putative) Inhibition of Ligation (Adenylation) by #162535 (solute) relative to DMSO (solvent)_# 162535 Molarity Titration DMSO DMSO DMSO 3uM 3uM 3uM 1uM 1uM 1uM 0 20 40 60 80 100 120 140 160 180 # 0018A # 0033B # 0034B Molarity Titration: 100uM-30nM MeanSpecificsignal/DMSO_% DMSO 100uM 30uM 10uM 3uM 1uM 300nM 100nM 30nM # 162535 1ul Undiluted Prep #1_080802 0.25ul Undiluted Prep #1_0808020.5ul 50% Glycerol diluted prep 36% 67% 42% 90% 12% 48% 25,000,000 217,000,000 45,000,000 For N=3 predicted IC50 approximates to ~ 2uM For N=1 predicted IC50 ~1uM (1ul undiluted prep #1) Therefore repeated using 0.25ul undiluted prep #1 Thus, now N=4 for 0.25ul undiluted prep #1 Repeated !Repeated !
- 8. Variable background was a confounding factor when it came to signal comparison for the different titrants Nevertheless, evident IC50 was approximately 1uM This concurs with assays #34 & #35
- 9. % (Putative) Inhibition of Ligation (Adenylation) by #1698 (solute) relative to DMSO (solvent)_# 1698 Molarity Titration DMSO DMSO 100uM 100uM 30uM 30uM 10uM 10uM 3uM 3uM 1uM 1uM 300nM 300nM 100nM 100nM 30nM 30nM 0 20 40 60 80 100 120 # 0018B # 0035A MeanSpecificSignal/DMSO_% IC50 resides here # 1698 28, 000,000 190, 000,000 27% 53% 38% 61% For N=1 predicted IC50 approximates to 10uM For N=2 predicted IC50 lies somewhere between 10uM & 3uM #0035A = 0.25ul undiluted prep #1_080802 Therefore repeated using (similarly) 0.25ul undiluted prep #1 Thus, for 0.25ul undiluted prep #1, now N=4 ! Repeated !Repeated !
- 10. 108, 000, 000 counts108, 000, 000 counts 153, 000, 000 counts153, 000, 000 counts
- 11. % (Putative) Inhibition of Ligation (Adenylation) by #45609 (solute) relative to DMSO (solvent)_# 45609 Molarity Titration DMSO DMSO 100uM 100uM 30uM30uM 10uM 10uM 3uM 3uM 1uM 1uM 300nM 300nM 100nM 100nM 30nM 30nM 0 20 40 60 80 100 120 140 # 0035B # 0016B Molar Titration MeanSpecificSignal_%DMSO IC50 resides here 199,000,000 counts ! 10,000,000 counts ! 0.9% 1.6% 7.9% 36% 68% IC50 resides here 12% 16% 22% 38% 74% Thus, for N=3 predicted of IC50 ~3-1uM However, above this threshold, titrants for respective assays differ considerably with respect to putative inhibition Assay #0016B = 0.5ul 50 % glycerol diluted prep Assay #0035B = 0.25ul undiluted prep #1_080802 Repeat ??Repeat ??
- 12. % (Putative) Inhibition of REL 1 Ligation (Adenylation) by #117079 (solute) relative to DMSO (solvent)_# 117079 Molarity Titration DMSO DMSO 100uM 100uM 30uM 30uM 10uM 10uM 3uM 3uM 1uM 1uM 300nM 300nM 100nM 100nM 30nM 30nM 0.0 20.0 40.0 60.0 80.0 100.0 120.0 140.0 BAC Prep #3_050203 BAC Prep #1_080802 #117079 Molarity Titration: 100uM-30nM SpecificSignal/DMSO_% 18.6%18.6% 54%54% 23.4%23.4% 75.6%75.6% 1ul Enzyme1ul Enzyme 0.25ul Enzyme0.25ul Enzyme Predicted IC50 Prep #3 ~30uM Predicted IC50 for prep #100uM-30uM The latter concurs with previous assays50S50S-1-1 100S100S-1-1
- 13. IC50 resides somewhere between 100uM & 30uM In the main, this concurs with previous assays, i.e.: #0011 & #0012 #0029B_Prep #1_080802 However, predicted IC50 for #29B_prep #3 approx. to 30uM rather than 100uM To illustrate discrepancies further: Putative inhibition @100uM for assays #0011 & #0012 >99% For assay #0029B ~ 80% For assay #0032A/B ~ 50 -70% # 117079 Molarity Titration: 1mM-1uM# 117079 Molarity Titration: 1mM-1uM 10S10S-1-1 10-20S10-20S-1-132A 32B5S5S-1-1 10S10S-1-1 1ul Prep #31ul Prep #3
- 14. % (Putative) Inhibition of REL 1 Ligation (Adenylation) by #125908 (solute) relative to DMSO (solvent): # 125908 Molarity Titration. Assay #0031A 1mM 1mM DMSO DMSO 300uM 100uM 100uM 30uM 30uM 10uM 10uM 3uM 3uM 1uM 1uM 0 20 40 60 80 100 120 140 # 31A_1 # 31A_2 Molarity Titration: 1mM-1uM SpecificSignal/DMSO_% 32.5% 77.7% 17.4% 48.4% 86.4% IC50 resides here IC50 resides here
- 15. % (Putative) Inhibition of REL 1 Ligation (Adenylation) by #125908 (solute) relative to DMSO (solvent): # 125908 Molarity Titration. Assay #0031B DMSO DMSO 1mM1mM 300uM 300uM 100uM 100uM 30uM 30uM 10uM 10uM 3uM 3uM 1uM 1uM 0 20 40 60 80 100 120 140 # 31B_1 # 31B_2 Molarity Titration: 1mM-1uM SpecificSignal/DMSO_% 17.4% 35.5% 60.9% 13.4% 30.2% 51.4% IC50 resides here IC50 resides here Predicted IC50 ~30uM: N=1 Predicted IC50 100uM-30uM: N=3 The latter concurs with previous assays
- 16. % (Putative) Inhibition of REL 1 Ligation (Adenylation) by #117079 (solute) relative to DMSO (solvent): #117079 Molarity Titration. Assay #0032A/B DMSO 1m M 300uM 100uM 30uM 10uM 3uM 1uM 0 20 40 60 80 100 120 N = 3 Molarity Titration #117079: 1mM - 1uM MeanSpecificSignal/DMSO_% DMSO 1mM 300uM 100uM 30uM 10uM 3uM 1uM Plan of action:Plan of action: 1. Either combine this data with #0029B_prep #1 (N=4) 2. Or repeat assay 0.25ul undiluted prep #1_080809
- 17. =10=10µMµM =100=100µMµM Low S:N precluded quantitation ! This was even true of depicted assay #0026, wherein cassette left for 6 days ! BAC 1 prep with low intrinsic activity (also common to H2L compounds)
- 18. Green = 1ul 50% Prep #3_050203 + 0.1% Triton X-100 Black = 1ul undiluted prep #1_080802 Blue = 1ul undiluted prep #1_080802 Green = 0.25ul undiluted prep #1_080802 200S200S-1-1 1000S1000S-1-133A 33B100S100S-1-1 1000S1000S-1-1
- 19. Loss of Specific Signal (DMSO) with Freeze thawing: REL1 'prep #3_050203' vs 'Prep #1_080802' #0031 #0032 #0027 #0027 #0029 #0029 #0034 #0035 #0036 0.00E+00 5.00E+07 1.00E+08 1.50E+08 2.00E+08 2.50E+08 DMSO_#1_0.25ul DMSO_#3_0.25_1ul Assays SpecificSignal #0027 #0029 #0031 #0032 #0034 #0035 #0036 0.25ul undiluted Prep #3_050203 0.25ul Prep #1_080802 1ul 50 % Prep #3_050203
- 20. SummarySummary 1. Mordant black has evident IC50 approximating to 1uM (N=5): Best lead !! 2. Regarding #45609, assays alight on IC50 approximating to 3-1uM (N=3) but differ with respect to putative titrant inhibition above that threshold; Repeat ? 3. With reference to #162535 & #1698, discrepancies evident between assay predicted IC50 values and for #1698 in particular N<3 for alternative values; There fore repeated (#0036) 4. Regarding #117079 & #125908, some assays display putative different IC50’s but an N>/=3 exists for one value 5. However, titrant values above the predicted IC50 thresholds also differ: Repeat ? In general, same enzyme batches concur more often and undiluted prep #1_080802 has yielded highest signal to noise Thus, in general and N>/= 3 derived from this REL 1 batch would constitute the ideal Future WorkFuture Work 1. Quantify #0036 2. Repeat other assays described to nullify IC50 and preferably titrant differences in general, with a bias towards N>/= 3 from the same enzyme batch 3. Submit putative inhibitory values to Graph Pad Prism© to compute exact IC50s’ 4. Complete titrations for #45201
