sp2016ren
•Download as PPTX, PDF•
0 likes•42 views
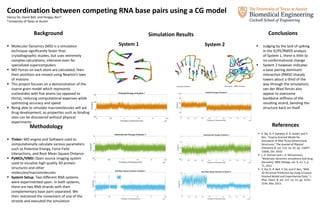
This document summarizes a study that uses coarse-grained molecular dynamics (MD) simulations to model the coordination between competing RNA base pairs. The study uses two RNA systems, each with two separated complementary strands, to test the coarse-grained model. For the first system, the simulation showed little conformational change, but the second system indicated a base pairing interaction as the RMSD sharply decreased partway through the simulation, suggesting the strands bent back together. The coarse-grained model reduces computational expenses compared to all-atom MD while optimizing accuracy and speed for simulating macromolecules like RNA, which could aid drug development efforts.
Report
Share
Report
Share
Recommended
Recommended
Predicting Functional Regions in Genomic DNA Sequences Using Artificial Neur...

Predicting Functional Regions in Genomic DNA Sequences Using Artificial Neur...International Journal of Engineering Inventions www.ijeijournal.com
During the process of molecular structure elucidation the selection of the most probable structural hypothesis may be based on chemical shift prediction. The prediction is carried out using either empirical or quantum-mechanical (QM) methods. When QM methods are used, NMR prediction commonly utilizes the GIAO option of the DFT approximation. In this approach the structural hypotheses are expected to be investigated by scientist. In this article we hope to show that the most rational manner by which to create structural hypotheses is actually by the application of an expert system capable of deducing all potential structures consistent with the experimental spectral data and specifically using 2D NMR data. When an expert system is used the best structure(s) can be distinguished using chemical shift prediction, which is best performed either by an incremental or neural net algorithm. The time-consuming QM calculations can then be applied, if necessary, to one or more of the 'best' structures to confirm the suggested solution.A systematic approach for the generation and verification of structural hypot...

A systematic approach for the generation and verification of structural hypot...US Environmental Protection Agency (EPA), Center for Computational Toxicology and Exposure
More Related Content
What's hot
Predicting Functional Regions in Genomic DNA Sequences Using Artificial Neur...

Predicting Functional Regions in Genomic DNA Sequences Using Artificial Neur...International Journal of Engineering Inventions www.ijeijournal.com
What's hot (19)
Gene association networks - Large-scale integration of data and text

Gene association networks - Large-scale integration of data and text
A distance-based method for phylogenetic tree reconstruction using algebraic ...

A distance-based method for phylogenetic tree reconstruction using algebraic ...
MODELING DEHYDRATION OF ORGANIC COMPOUNDS BY MEANS OF POLYMER MEMBRANES WITH ...

MODELING DEHYDRATION OF ORGANIC COMPOUNDS BY MEANS OF POLYMER MEMBRANES WITH ...
International Journal of Engineering Research and Development (IJERD)

International Journal of Engineering Research and Development (IJERD)
Gene association networks: Large-scale integration of data and text

Gene association networks: Large-scale integration of data and text
Gene association networks: Large-scale integration of data and text

Gene association networks: Large-scale integration of data and text
Gene association networks: Large-scale integration of data and text

Gene association networks: Large-scale integration of data and text
Predicting Functional Regions in Genomic DNA Sequences Using Artificial Neur...

Predicting Functional Regions in Genomic DNA Sequences Using Artificial Neur...
Similar to sp2016ren
During the process of molecular structure elucidation the selection of the most probable structural hypothesis may be based on chemical shift prediction. The prediction is carried out using either empirical or quantum-mechanical (QM) methods. When QM methods are used, NMR prediction commonly utilizes the GIAO option of the DFT approximation. In this approach the structural hypotheses are expected to be investigated by scientist. In this article we hope to show that the most rational manner by which to create structural hypotheses is actually by the application of an expert system capable of deducing all potential structures consistent with the experimental spectral data and specifically using 2D NMR data. When an expert system is used the best structure(s) can be distinguished using chemical shift prediction, which is best performed either by an incremental or neural net algorithm. The time-consuming QM calculations can then be applied, if necessary, to one or more of the 'best' structures to confirm the suggested solution.A systematic approach for the generation and verification of structural hypot...

A systematic approach for the generation and verification of structural hypot...US Environmental Protection Agency (EPA), Center for Computational Toxicology and Exposure
The major objective of the conformational analysis is to gain insight into the conformational characteristic of flexible biomolecules and drugs and identify the relation between the role of conformational flexibility and their activity.
The significance of conformational analysis not just extends to computational docking and screening but also to lead optimization
Conformational analysis – Alignment of molecules in 3D QSAR

Conformational analysis – Alignment of molecules in 3D QSARNational Institute of Pharmaceutical Education and Research (NIPER), Hyderabad
In silico prediction of small molecule properties is widely used today in industry and academia. NMR spectra, in particular, are predicted by a variety of software packages. In this array of software options, two main approaches are used:
Database-based. Compounds are compared against a database, the result is calculated using data for close structural relatives found in the dataset.
Regression-based. An experimental database is used to calculate the parameters for non-linear regression. The chemical shift is calculated by a non-linear function of variables which describe characteristic features of the molecule of interest.
These two outlined approaches require different strategies for implementation and optimization. Database-based results are improved by acquiring larger databases and/or including user-specific data into the calculation. Non-linear regression algorithms can be improved through the regression itself, or by improving the structural descriptorsNMR Chemical Shift Prediction by Atomic Increment-Based Algorithms

NMR Chemical Shift Prediction by Atomic Increment-Based AlgorithmsUS Environmental Protection Agency (EPA), Center for Computational Toxicology and Exposure
Similar to sp2016ren (20)
A systematic approach for the generation and verification of structural hypot...

A systematic approach for the generation and verification of structural hypot...
Conformational analysis – Alignment of molecules in 3D QSAR

Conformational analysis – Alignment of molecules in 3D QSAR
NMR Chemical Shift Prediction by Atomic Increment-Based Algorithms

NMR Chemical Shift Prediction by Atomic Increment-Based Algorithms
SIMILARITY ANALYSIS OF DNA SEQUENCES BASED ON THE CHEMICAL PROPERTIES OF NUCL...

SIMILARITY ANALYSIS OF DNA SEQUENCES BASED ON THE CHEMICAL PROPERTIES OF NUCL...
SIMILARITY ANALYSIS OF DNA SEQUENCES BASED ON THE CHEMICAL PROPERTIES OF NUCL...

SIMILARITY ANALYSIS OF DNA SEQUENCES BASED ON THE CHEMICAL PROPERTIES OF NUCL...
The Assembly, Structure and Activation of Influenza a M2 Transmembrane Domain...

The Assembly, Structure and Activation of Influenza a M2 Transmembrane Domain...
sp2016ren
- 1. Coordination between competing RNA base pairs using a CG model Danny Vo, David Bell, and Pengyu Ren* *University of Texas at Austin Background Molecular Dynamics (MD) is a simulation technique significantly faster than crystallographic studies, but uses extremely complex calculations, intensive even for specialized supercomputers MD Forces on each atom are calculated, then their positions are moved using Newton’s laws of motions This project focuses on a demonstration of the coarse grain model which represents nucleotides with five atoms (as opposed to thirty), reducing computational expenses while optimizing accuracy and speed Being able to simulate macromolecules will aid drug development, as properties such as binding sites can be discovered without physical experiments Methodology Tinker: MD engine and Software used to computationally calculate various parameters such as Potential Energy, Force Field Interactions, and Root Mean Square Distance PyMOL/VMD: Open source imaging system used to visualize high quality 3D protein structures and other molecules/macromolecules System Setup: Two different RNA systems were experimented upon. In both systems, there are two RNA strands with their complementary base pairs separated. We then restrained the movement of one of the strands and executed the simulation Simulation Results Conclusions Judging by the lack of spiking in the IE/PE/RMDS analysis of System 1, there is little to no conformational change System 2 however indicates a base pairing dominant interaction (RMSD sharply lowers about a third of the way through the simulation). van der Waal forces also appear to overcome backbone stiffness of the resulting strand, bending the structure back on itself References Z. Xia, D. P. Gardner, R. R. Gutell, and P. Ren, “Coarse-Grained Model for Simulation of RNA Three-Dimensional Structures,” The Journal of Physical Chemistry B, vol. 114, no. 42, pp. 13497– 13506, Oct. 2010. J. D. Durrant and J. A. McCammon, “Molecular dynamics simulations and drug discovery,” BMC biology, vol. 9, no. 1, p. 71, 2011. Z. Xia, D. R. Bell, Y. Shi, and P. Ren, “RNA 3D Structure Prediction by Using a Coarse- Grained Model and Experimental Data,” J. Phys. Chem. B, vol. 117, no. 11, pp. 3135– 3144, Mar. 2013. System 1 System 2 Starting orientation Orientation ~7000th timestep