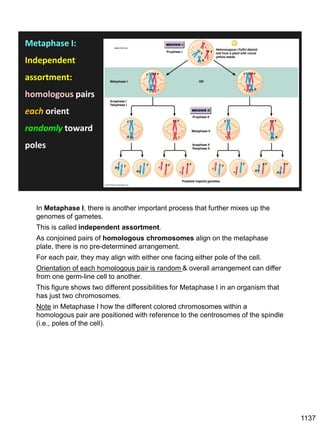
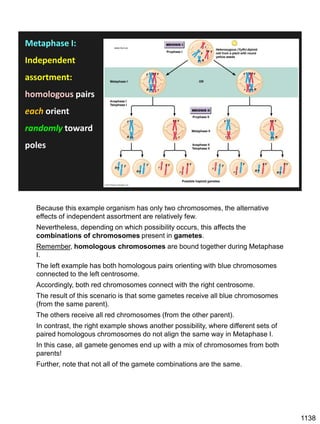
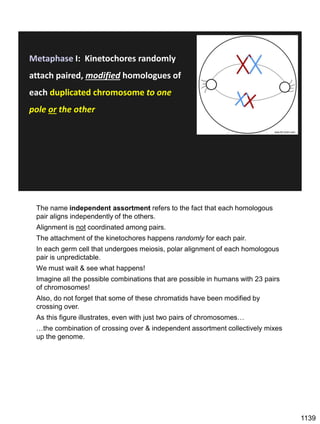
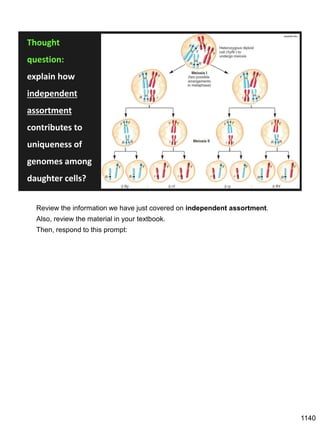
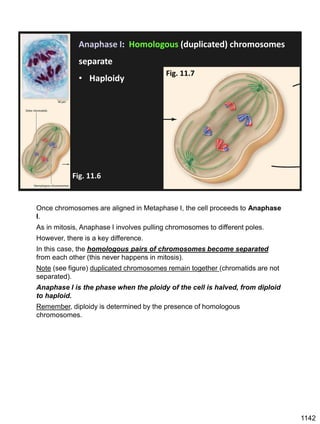
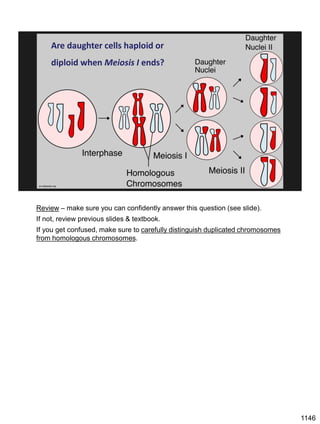
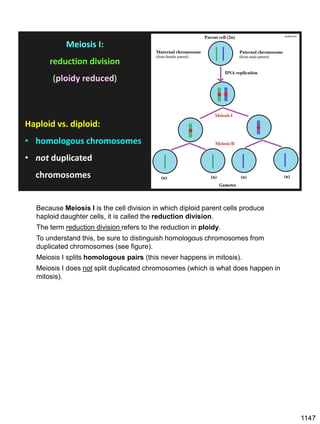
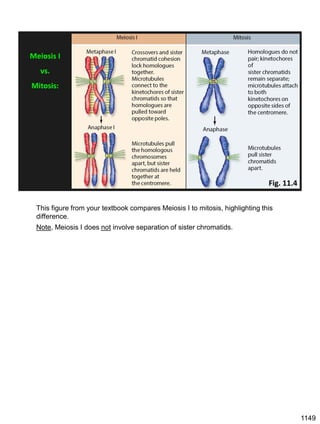
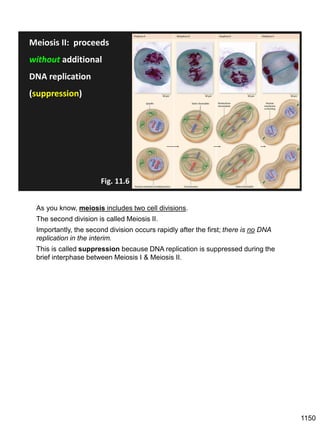
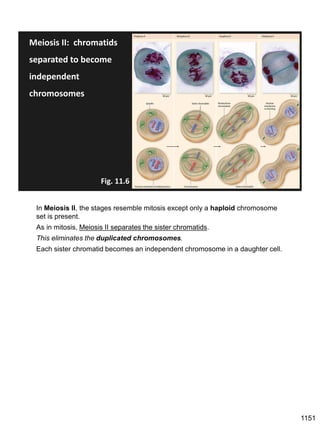
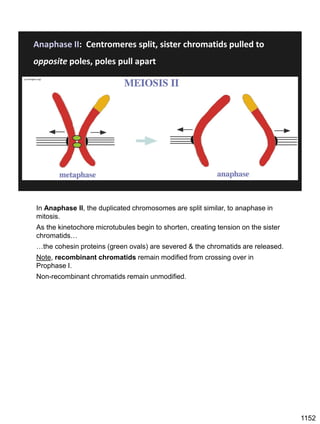
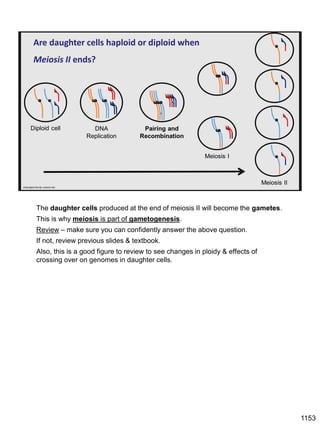
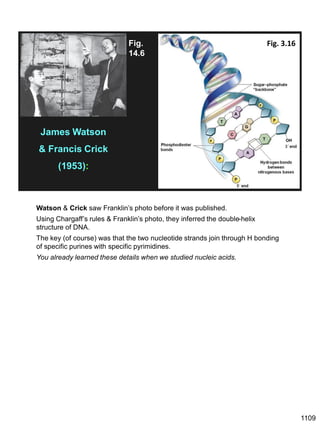
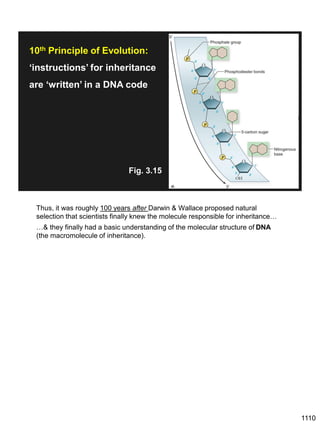
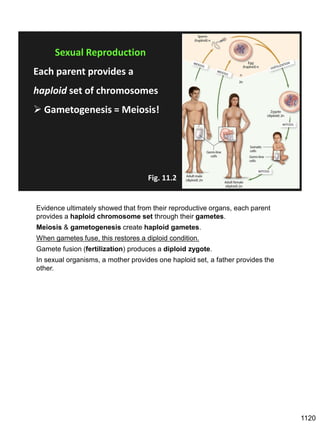
Erwin Chargaff discovered in 1950 that guanine always pairs with cytosine and adenine always pairs with thymine in DNA. He also found that different species have different ratios of these base pairs. In 1953, Rosalind Franklin used x-ray crystallography to take a photo of DNA structure. James Watson and Francis Crick used Chargaff's rules and Franklin's photo to deduce the double helix structure of DNA. This provided the first understanding of DNA's molecular structure and established it as the molecule responsible for inheritance.










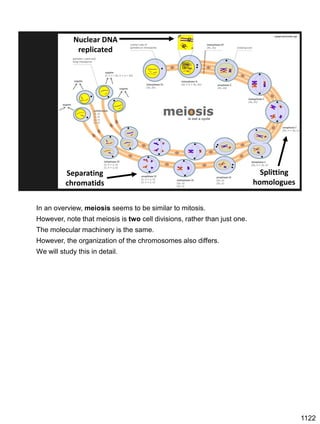
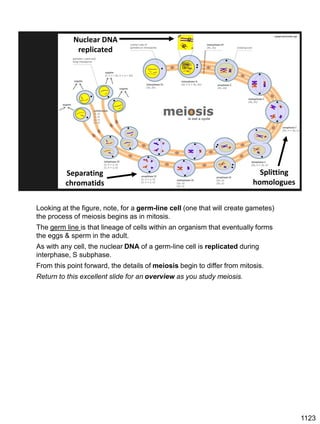
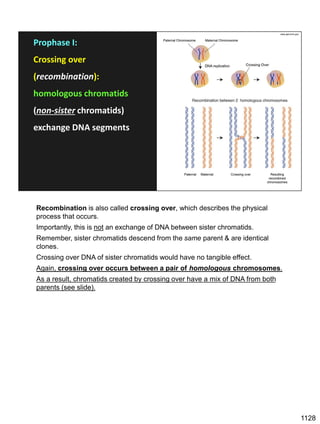
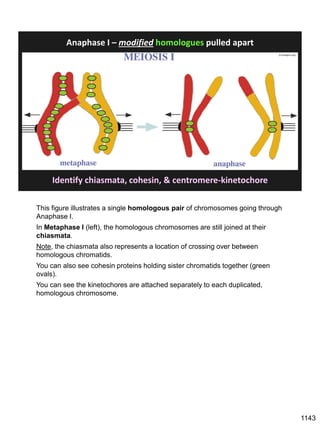
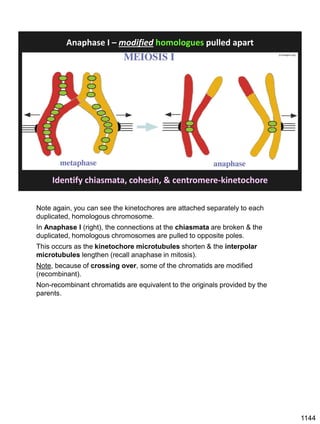
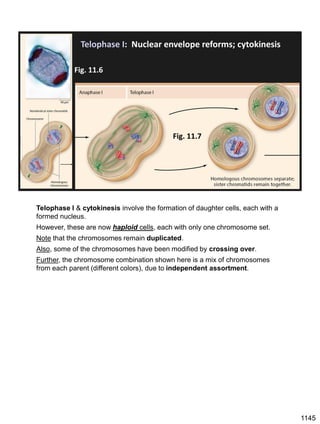
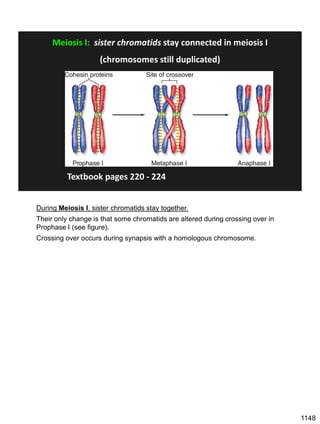
![Prophase I:
Recombination: homologous chromatids (non-sister chromatids)
exchange DNA segments
Page 221
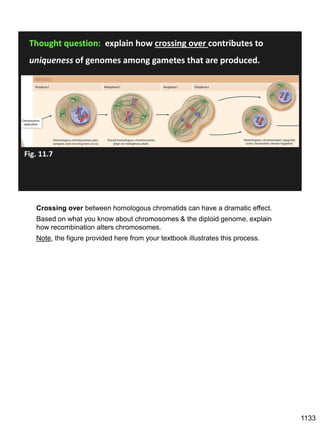
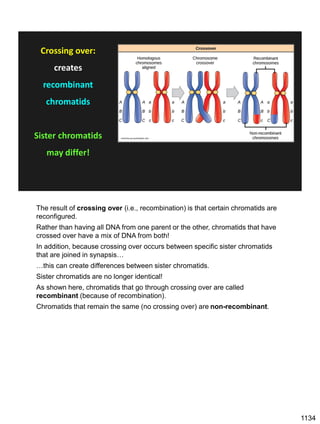
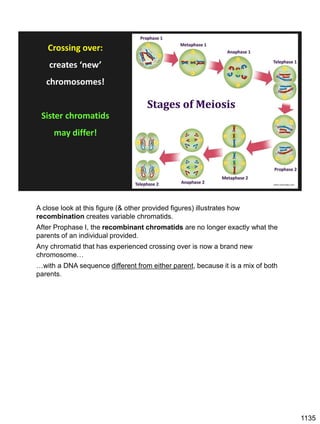
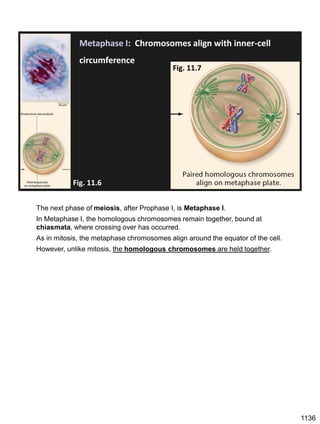
During Prophase I, homologous chromosomes can swap segments!
This is called recombination because it mixes the DNA of one haploid set with
that of the other.
In other words, DNA of different parents, which is normally kept separate,
becomes mixed.
[this refers to the parents of the individual whose cells are in meiosis]
Note, in your textbook (as in this figure), homologous chromosomes descending
from different parents are color coded.
One parent is shown as red, the other as blue.
1127](https://image.slidesharecdn.com/workshop22principlesfall2023notes1-231115214048-fa92c61d/85/Workshop22_Principles_Fall2023_Notes-1-pdf-21-320.jpg)