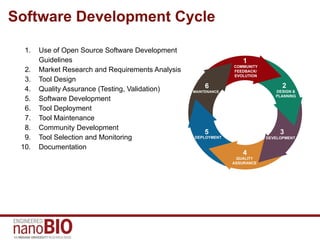
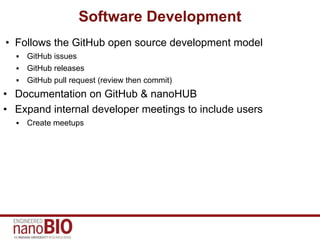
This document discusses the software development process and engineering practices used for tools created by the IU NanoBio group. It outlines the development cycle including requirements analysis, design, testing, deployment, maintenance, and community development. The group follows open source practices on GitHub for development and uses continuous integration, unit testing, and documentation on GitHub and nanoHUB. Quality assurance is conducted through detailed testing checklists. Tools and frameworks are on different release cycles, with tools receiving more rapid initial releases and maturing to longer cycles.