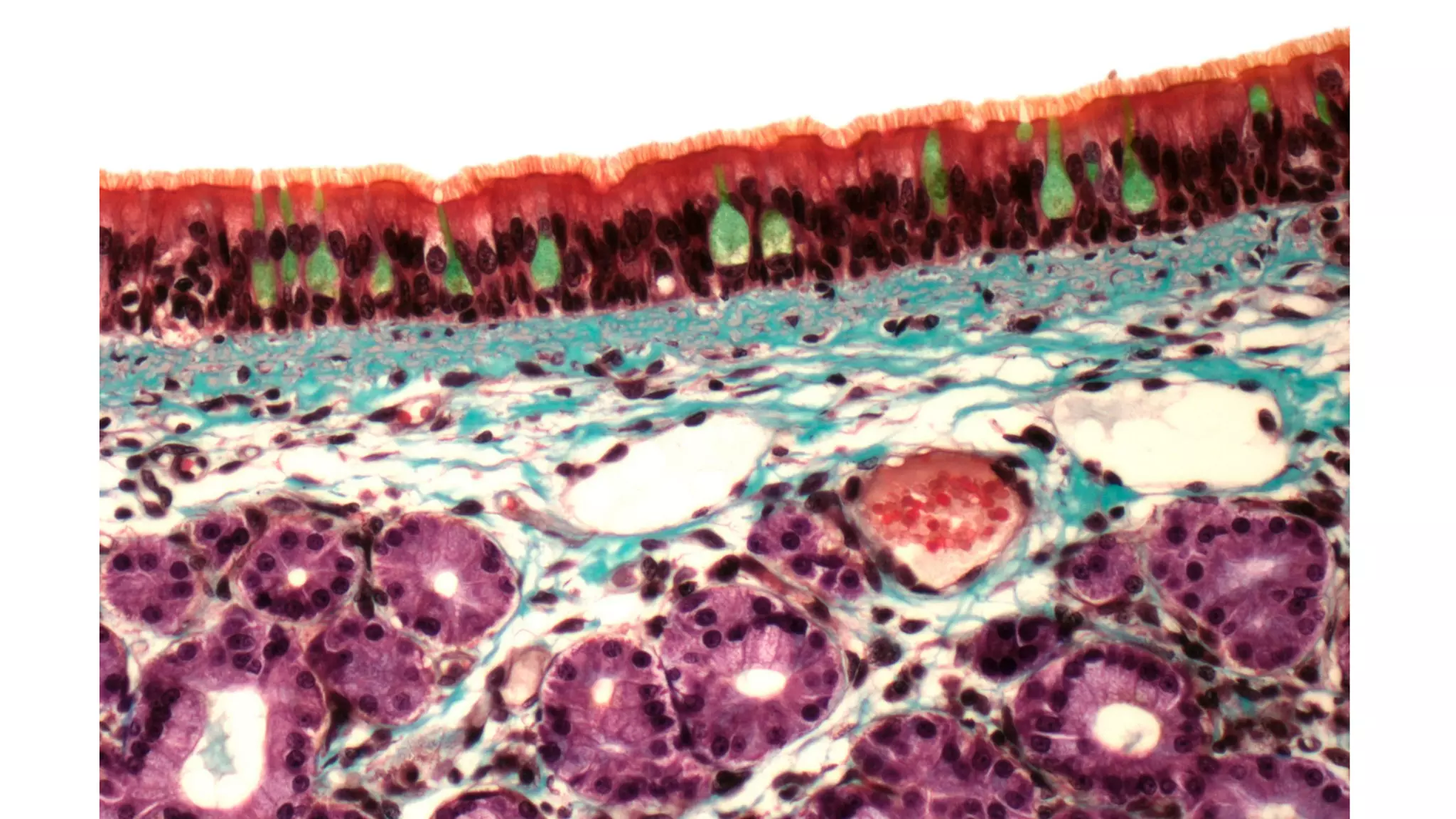
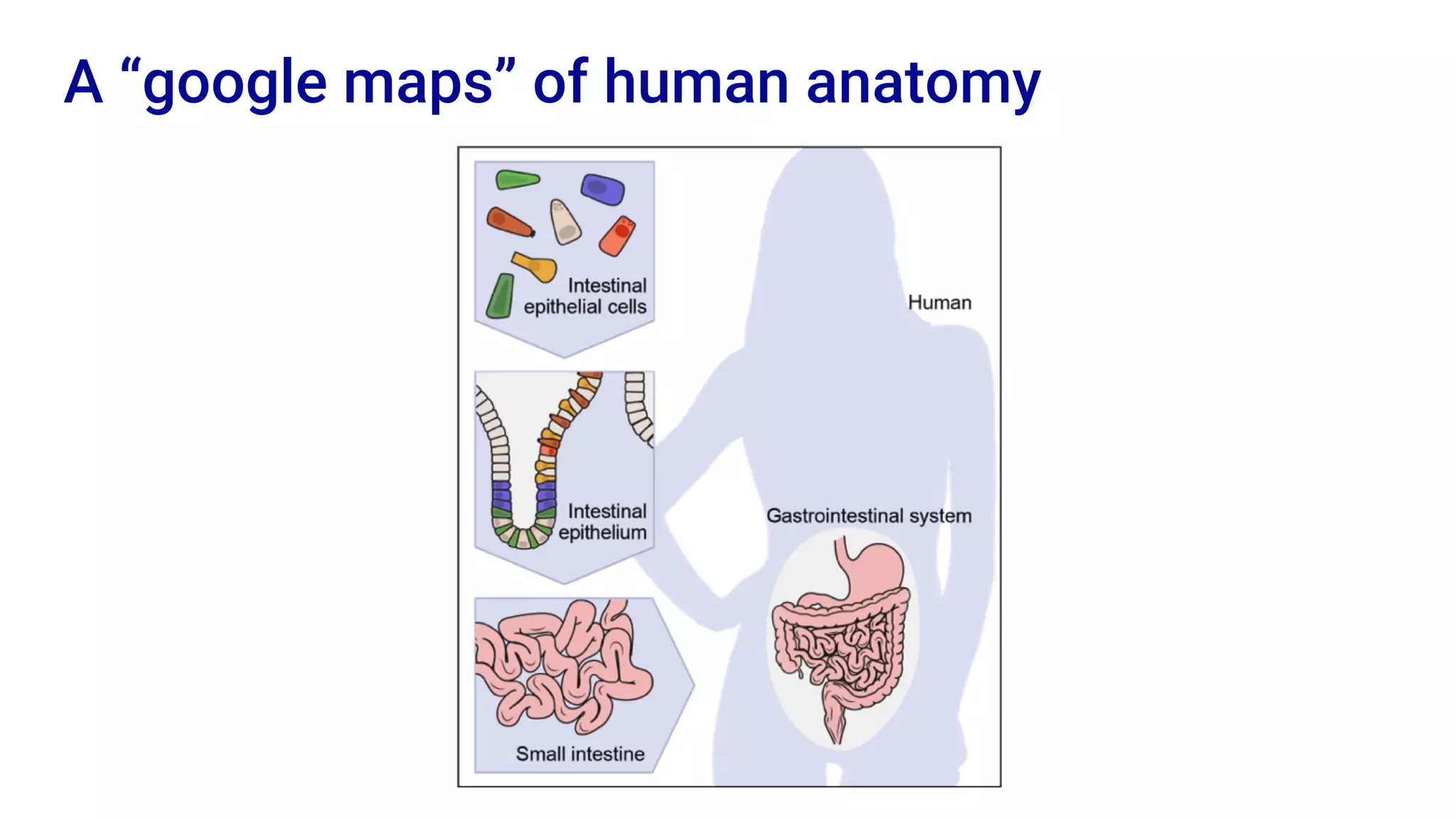
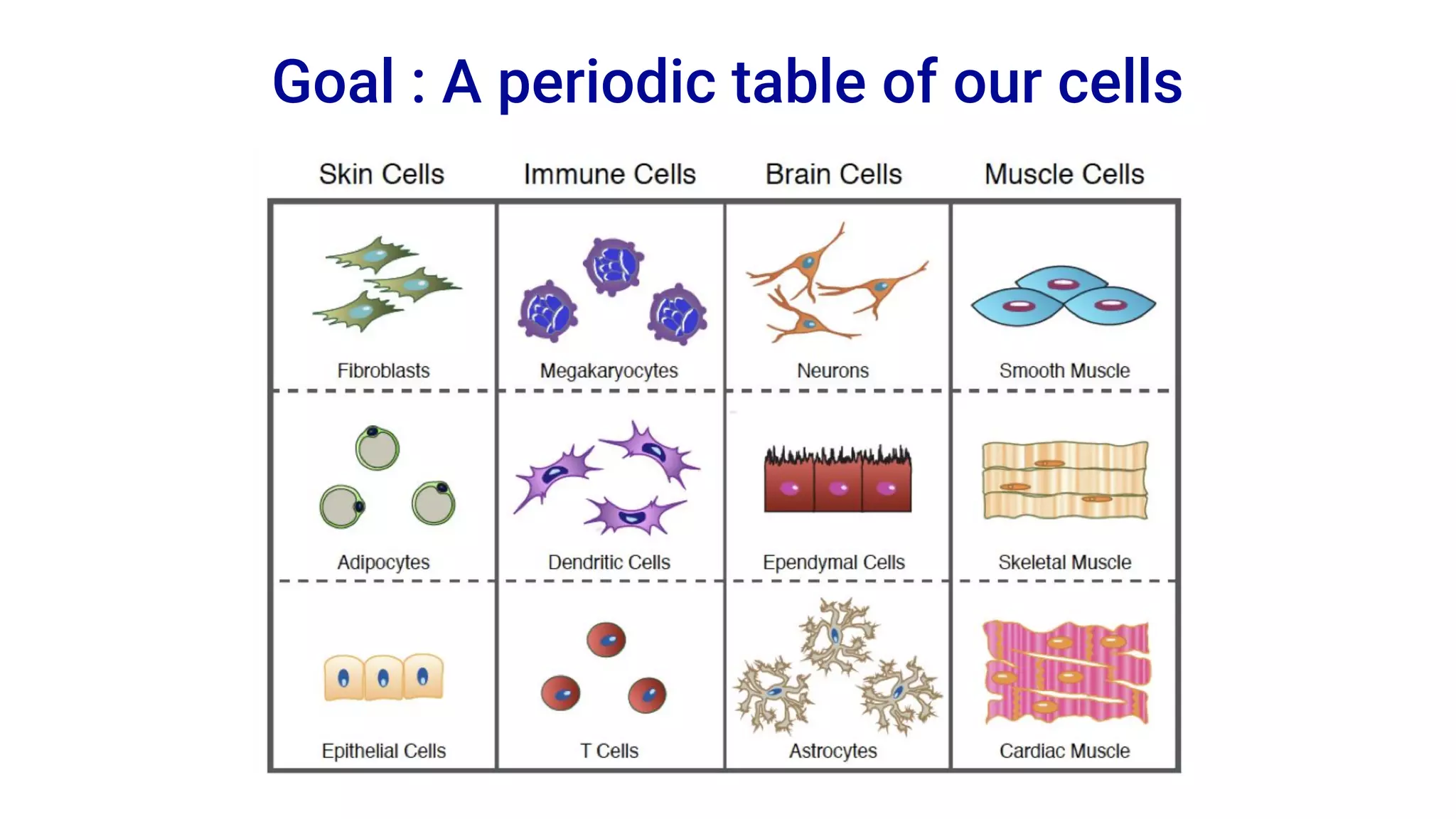
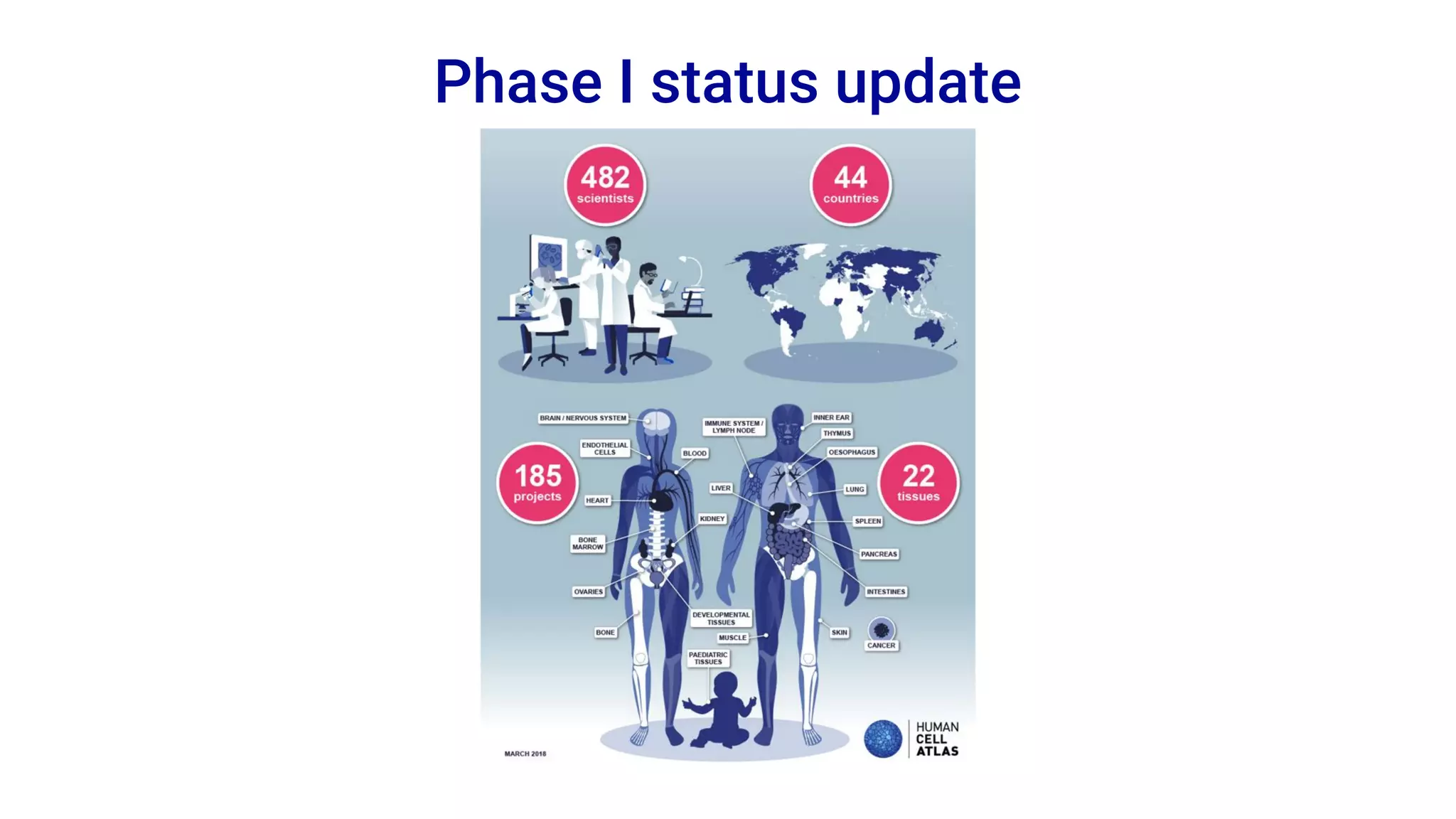
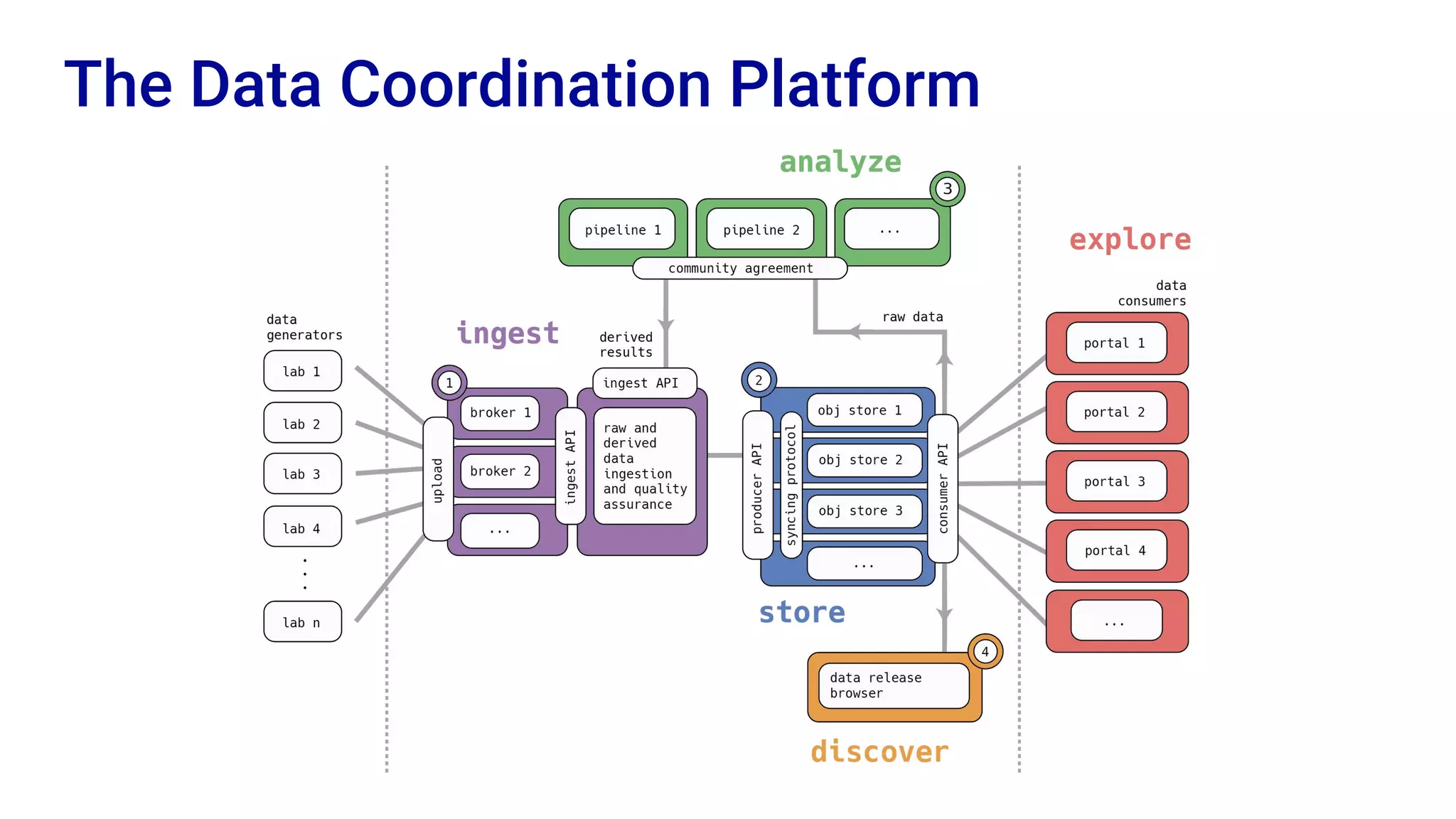
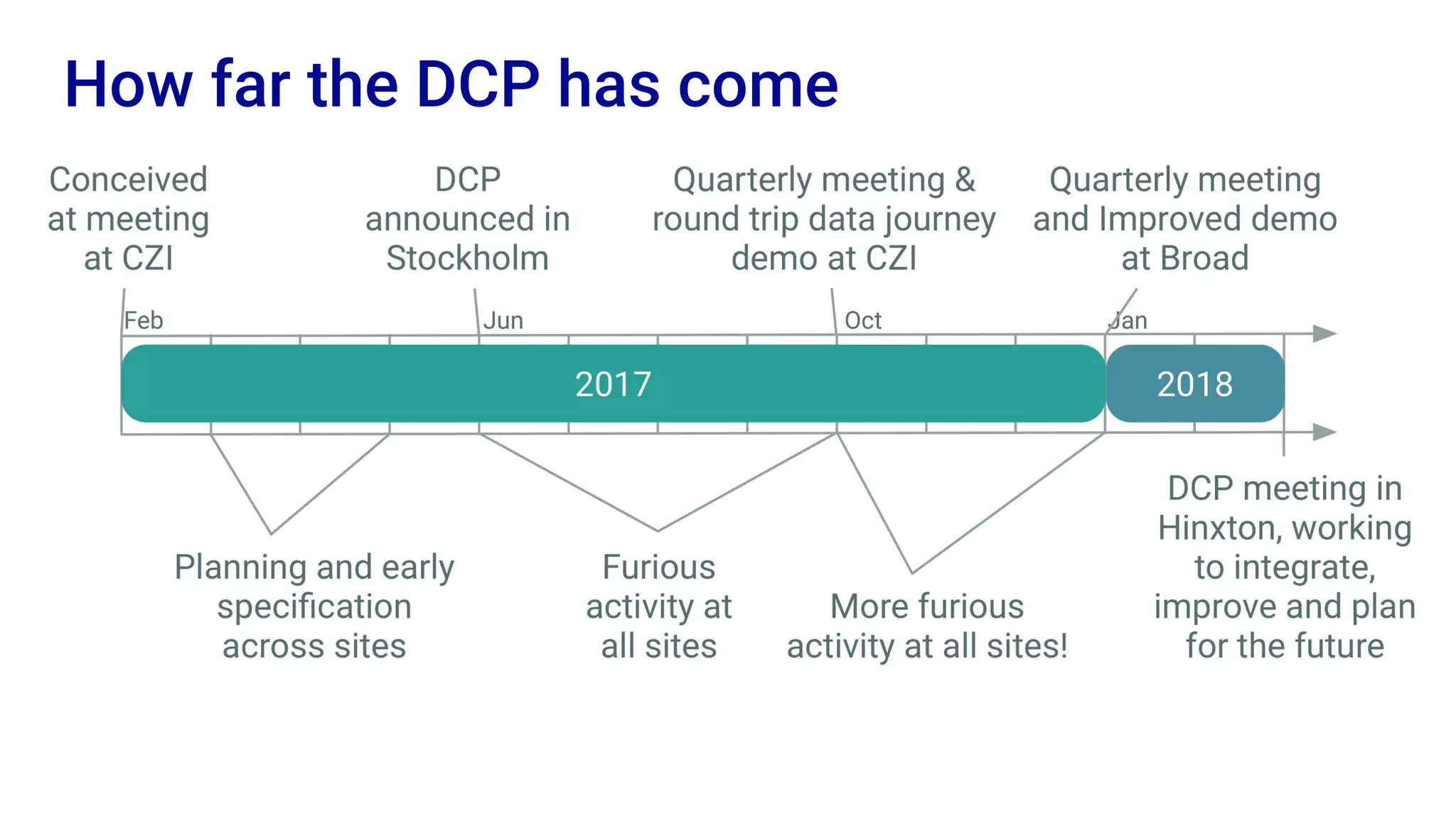
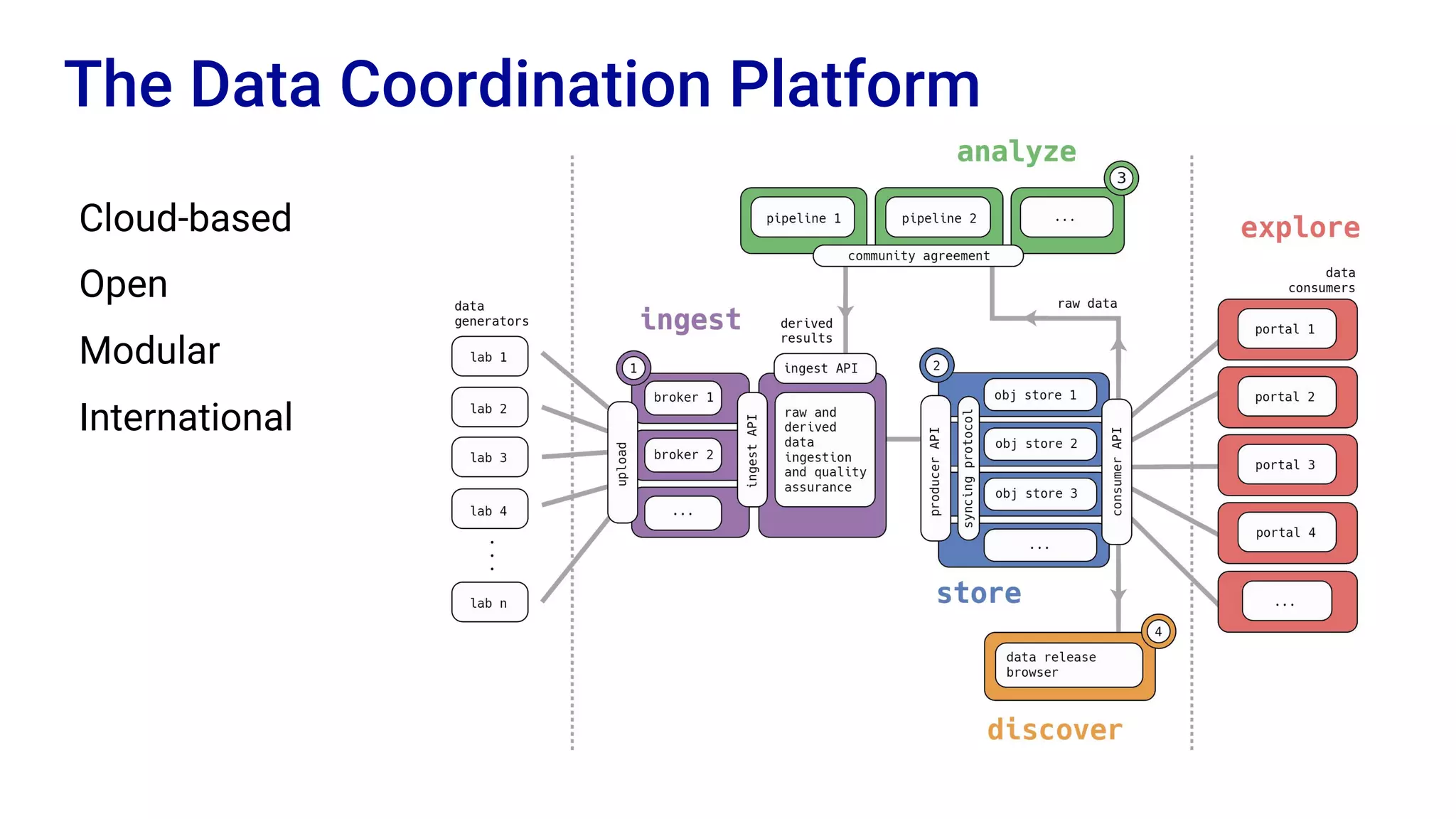
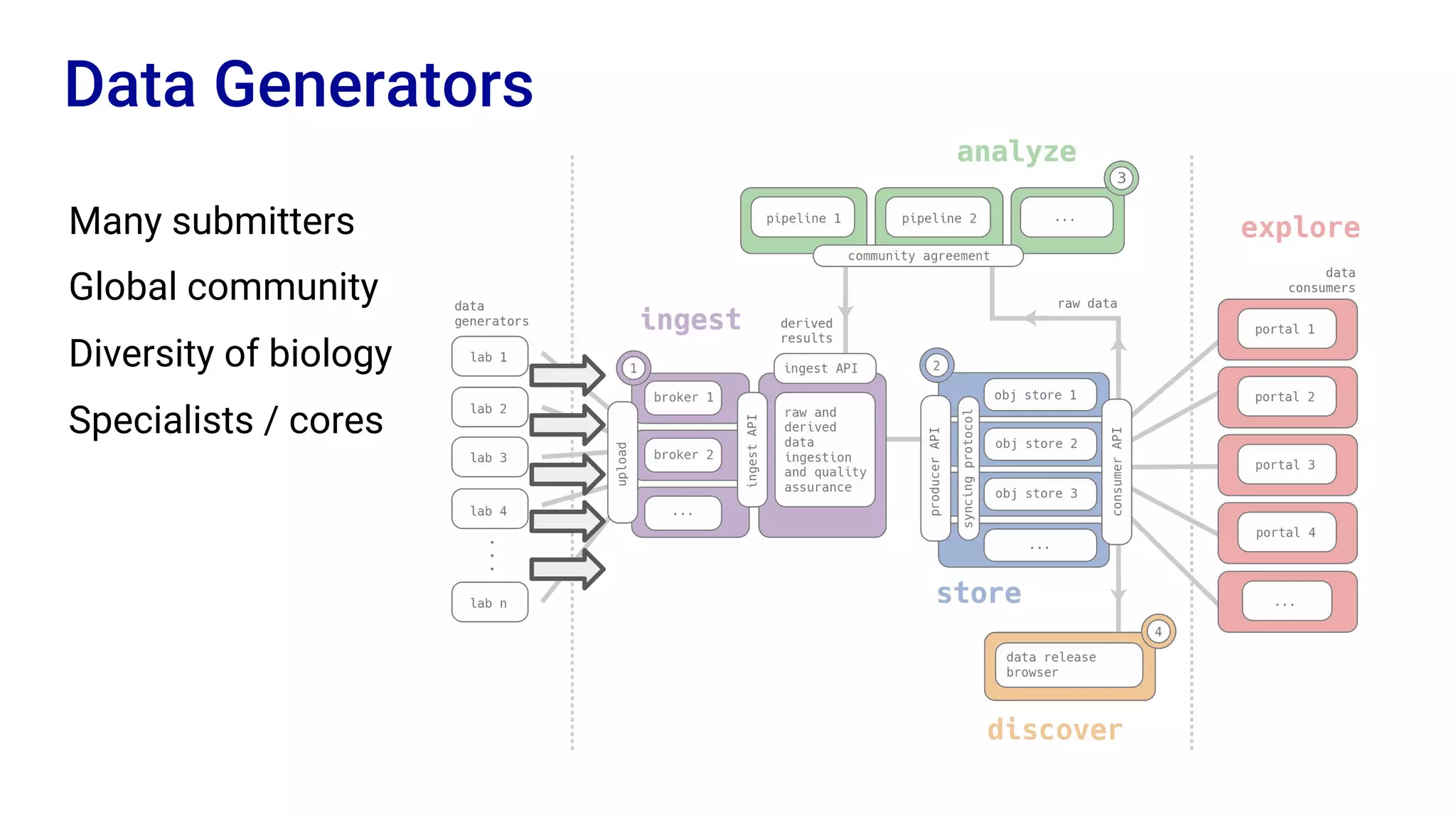
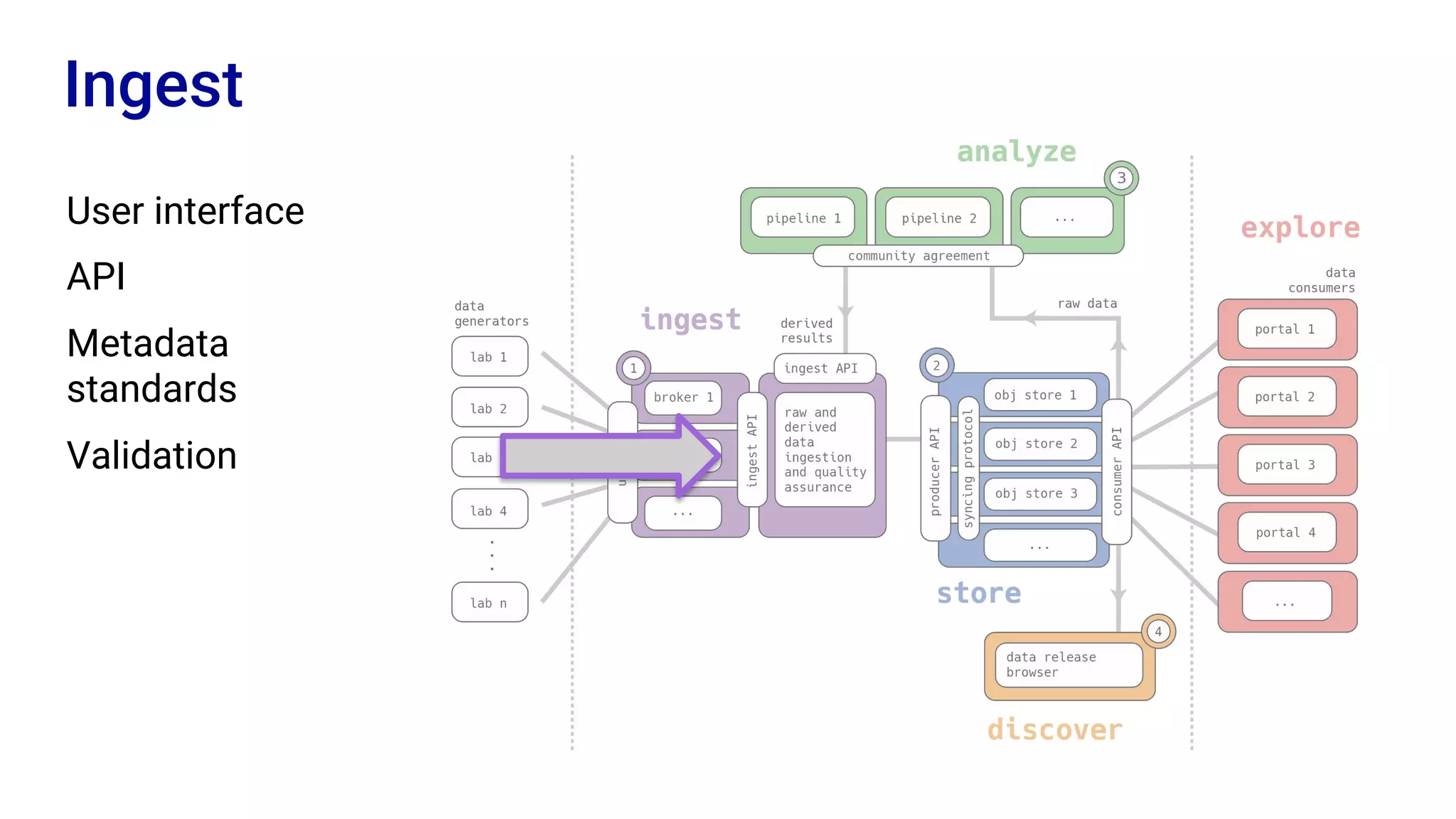
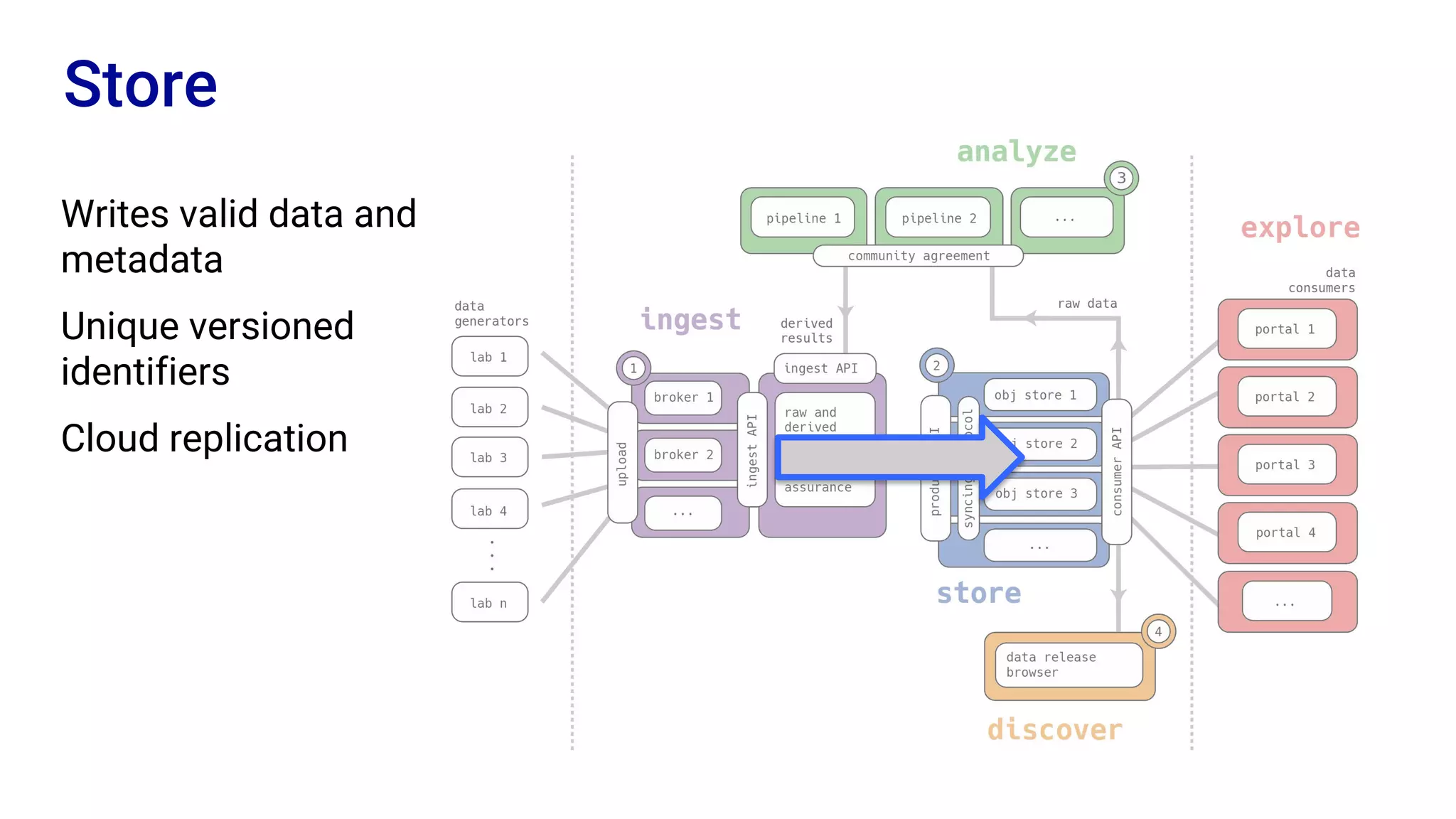
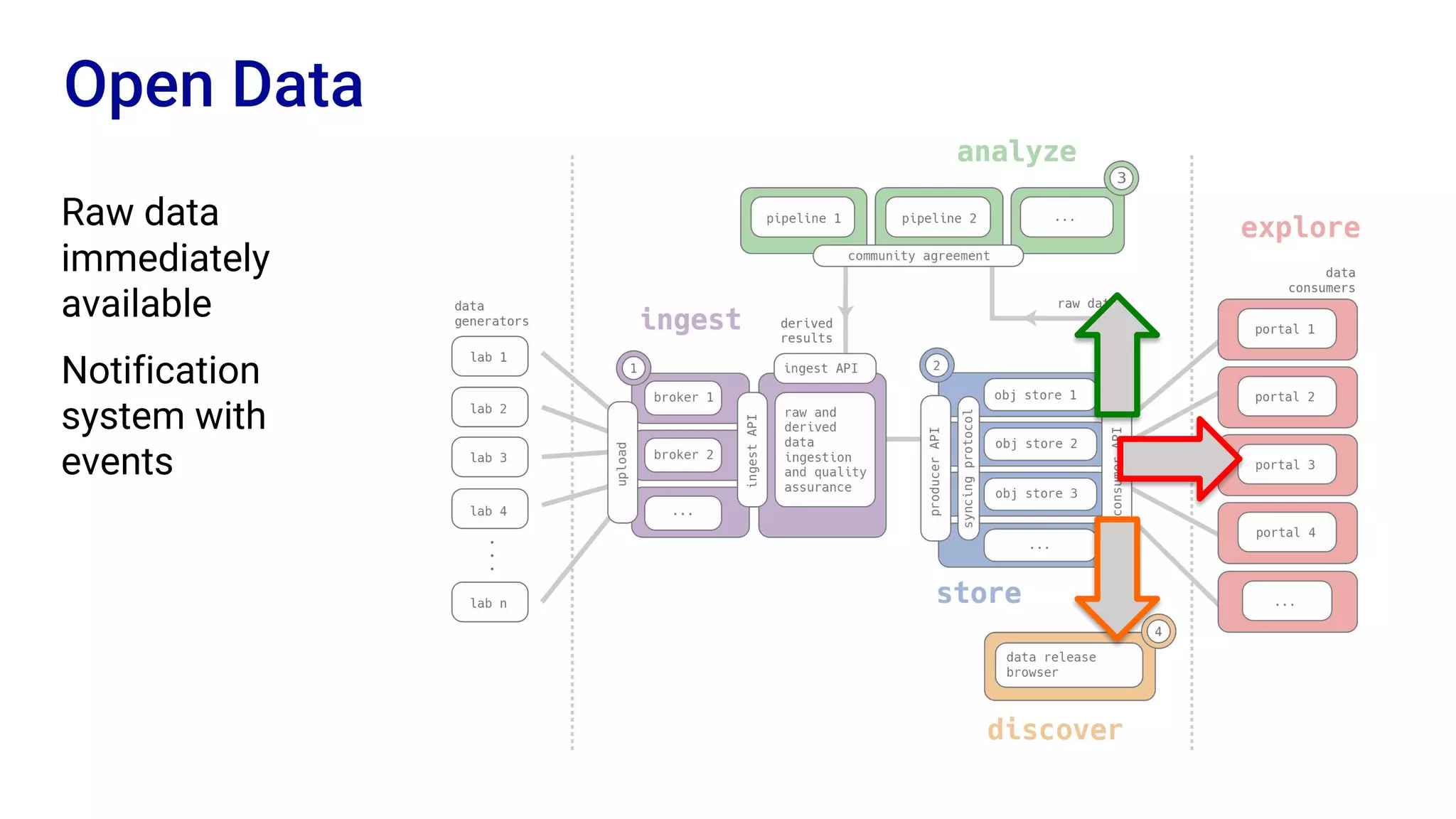
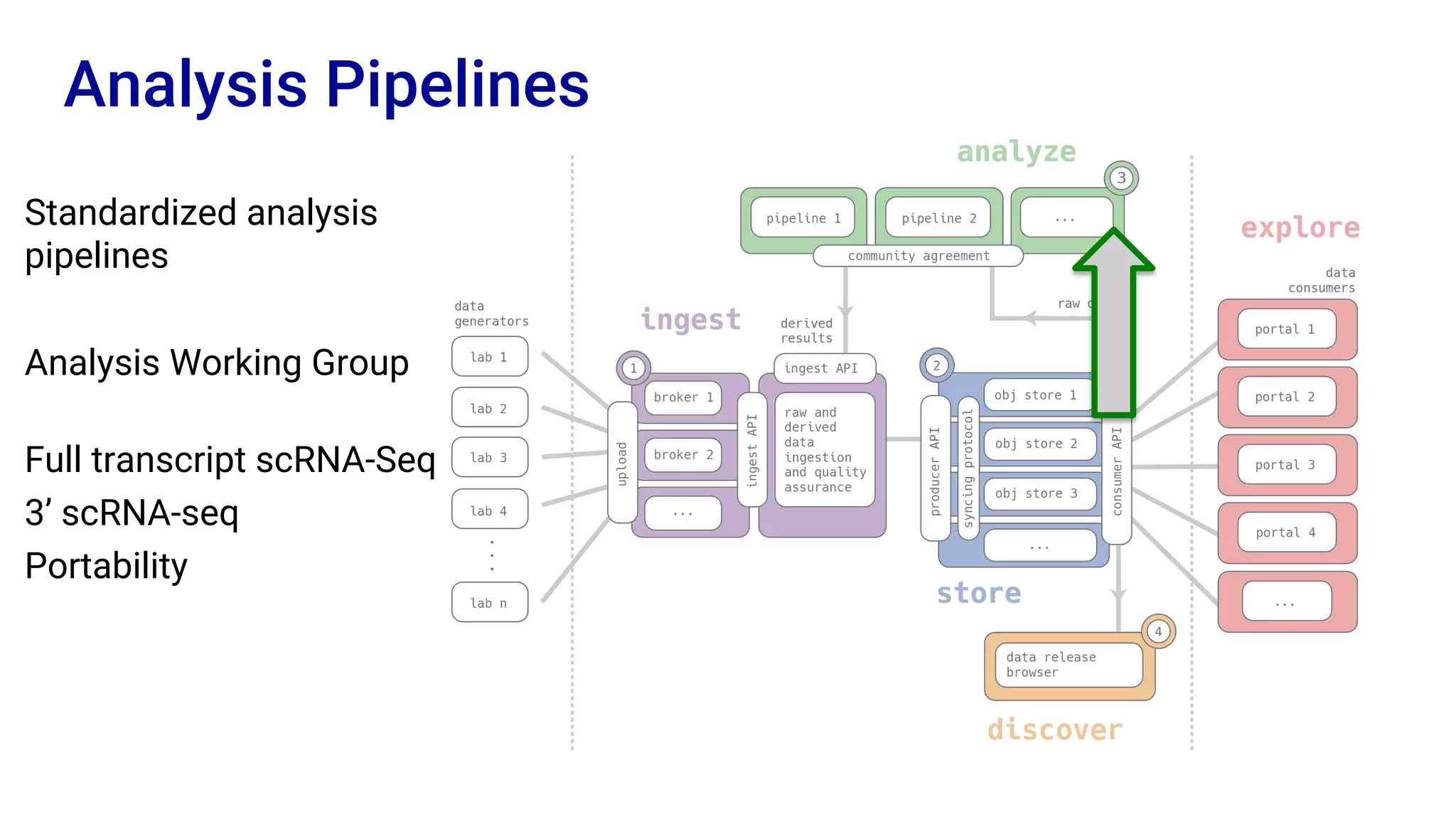
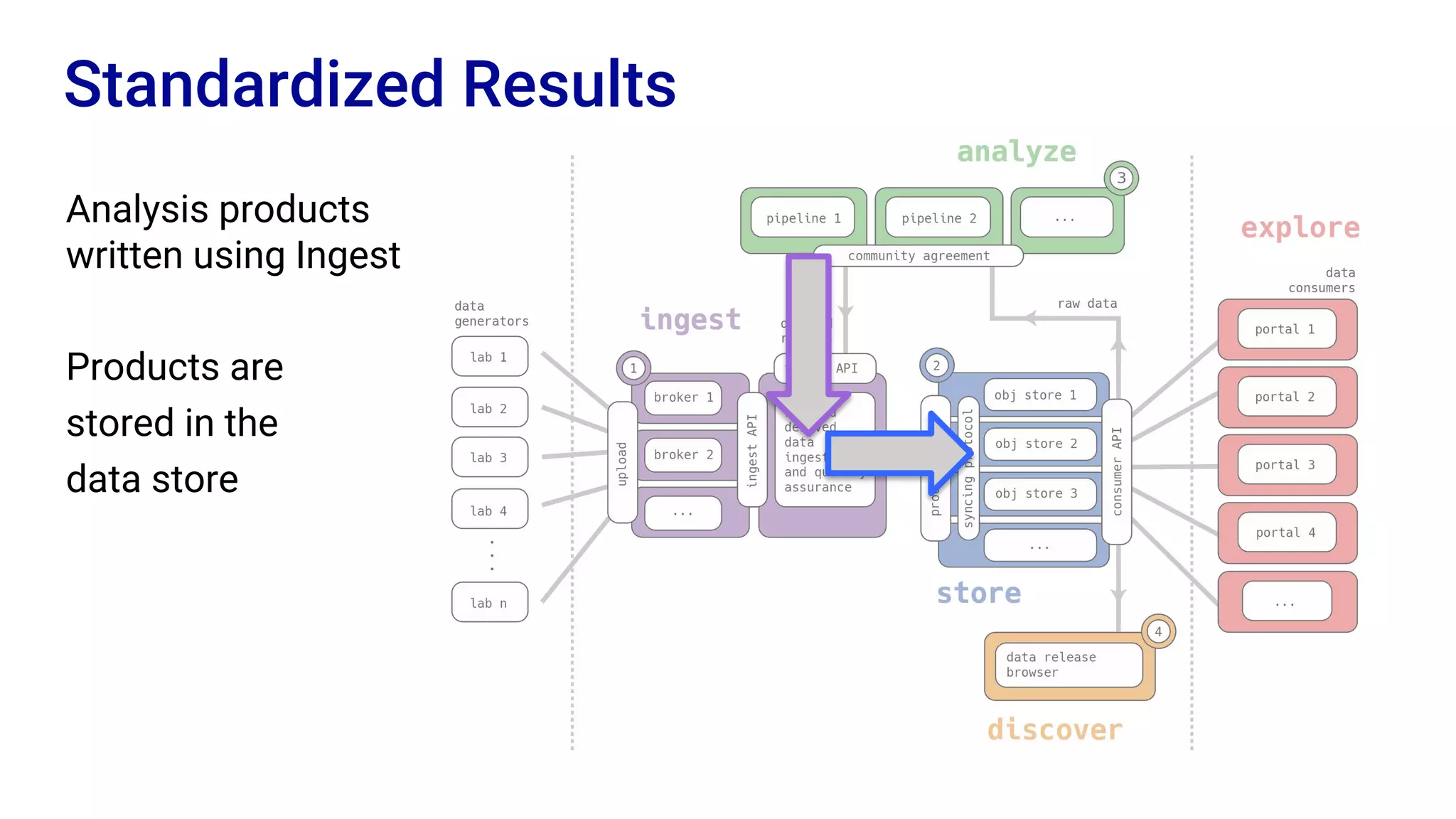
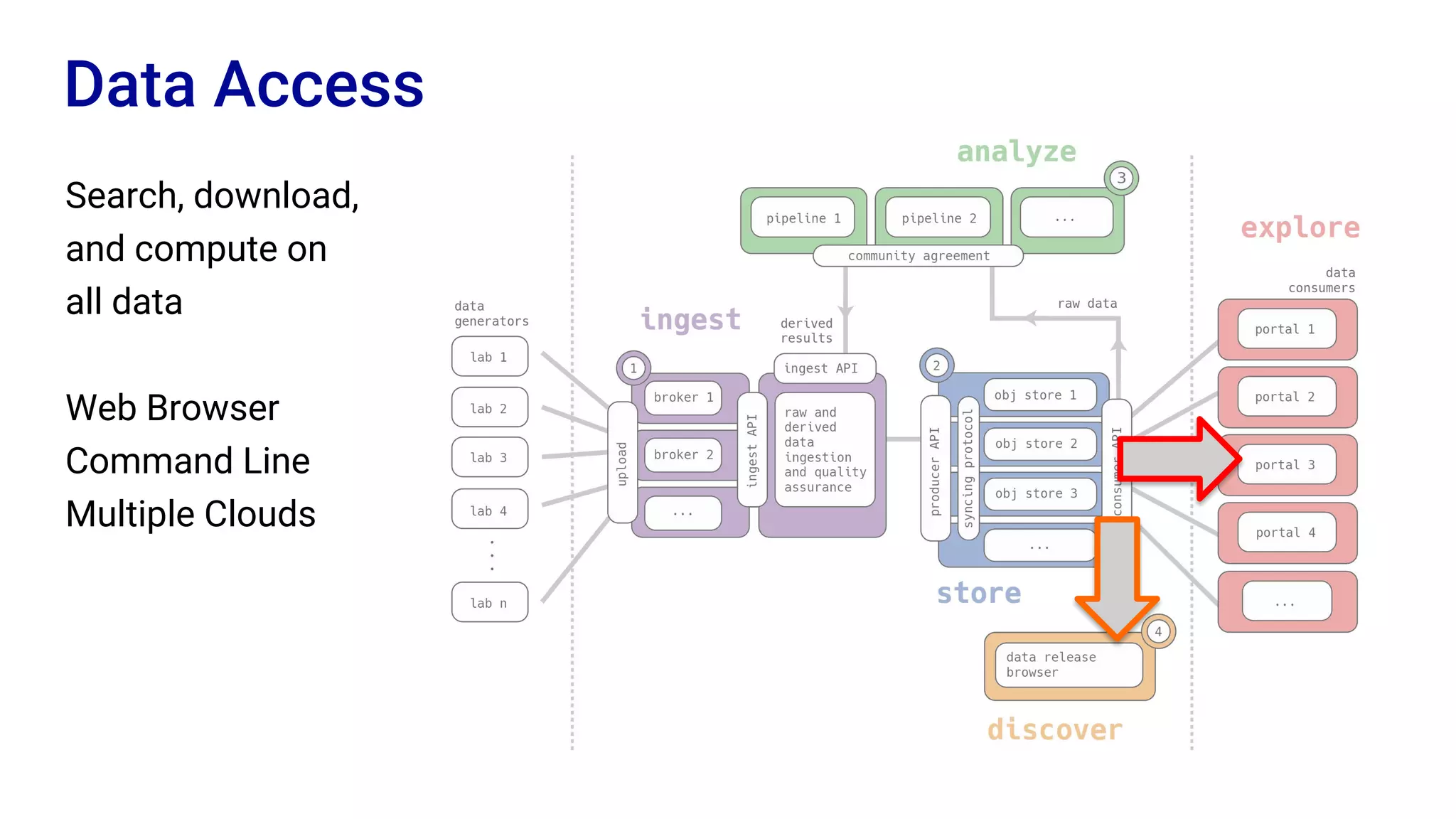
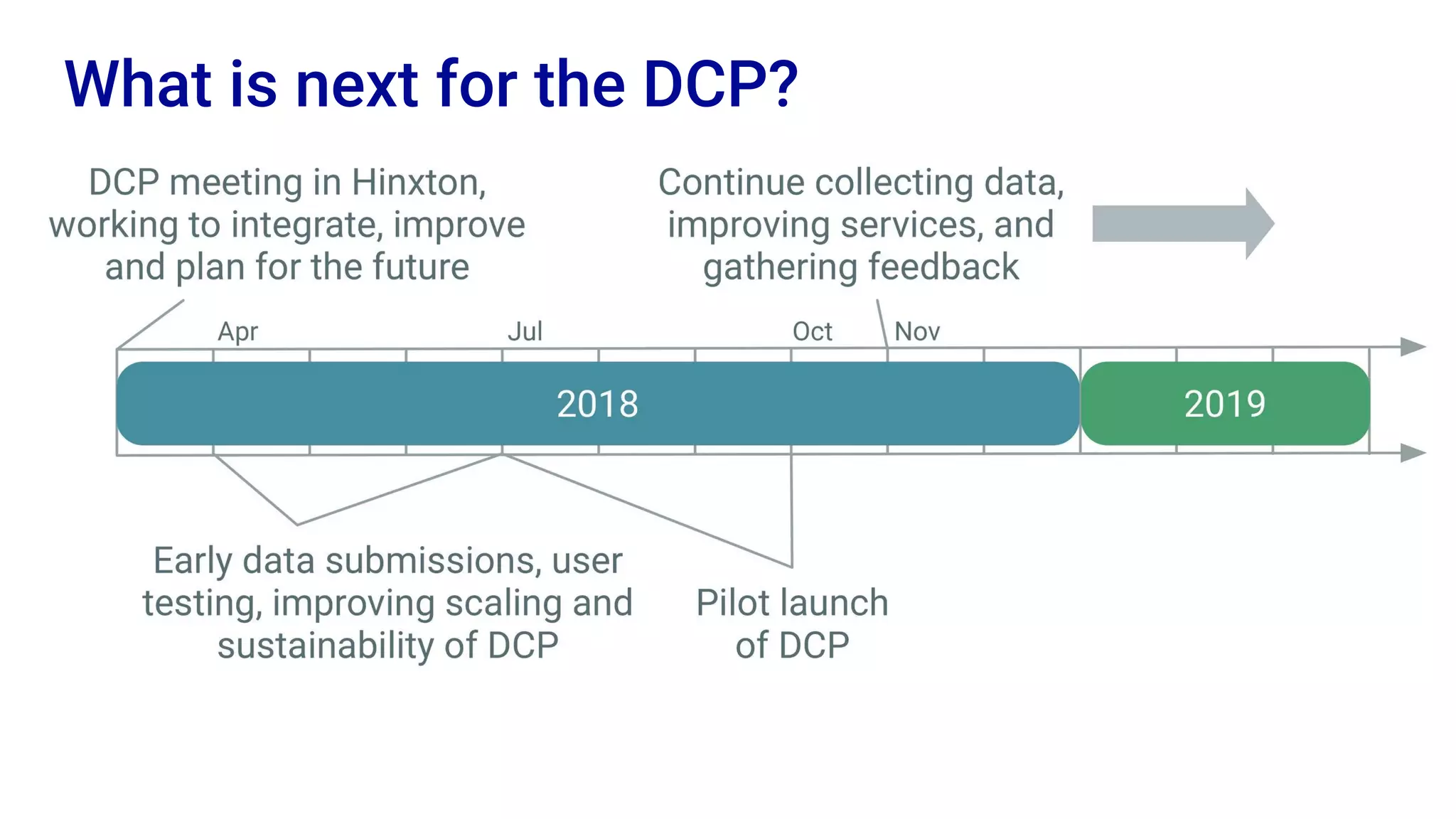
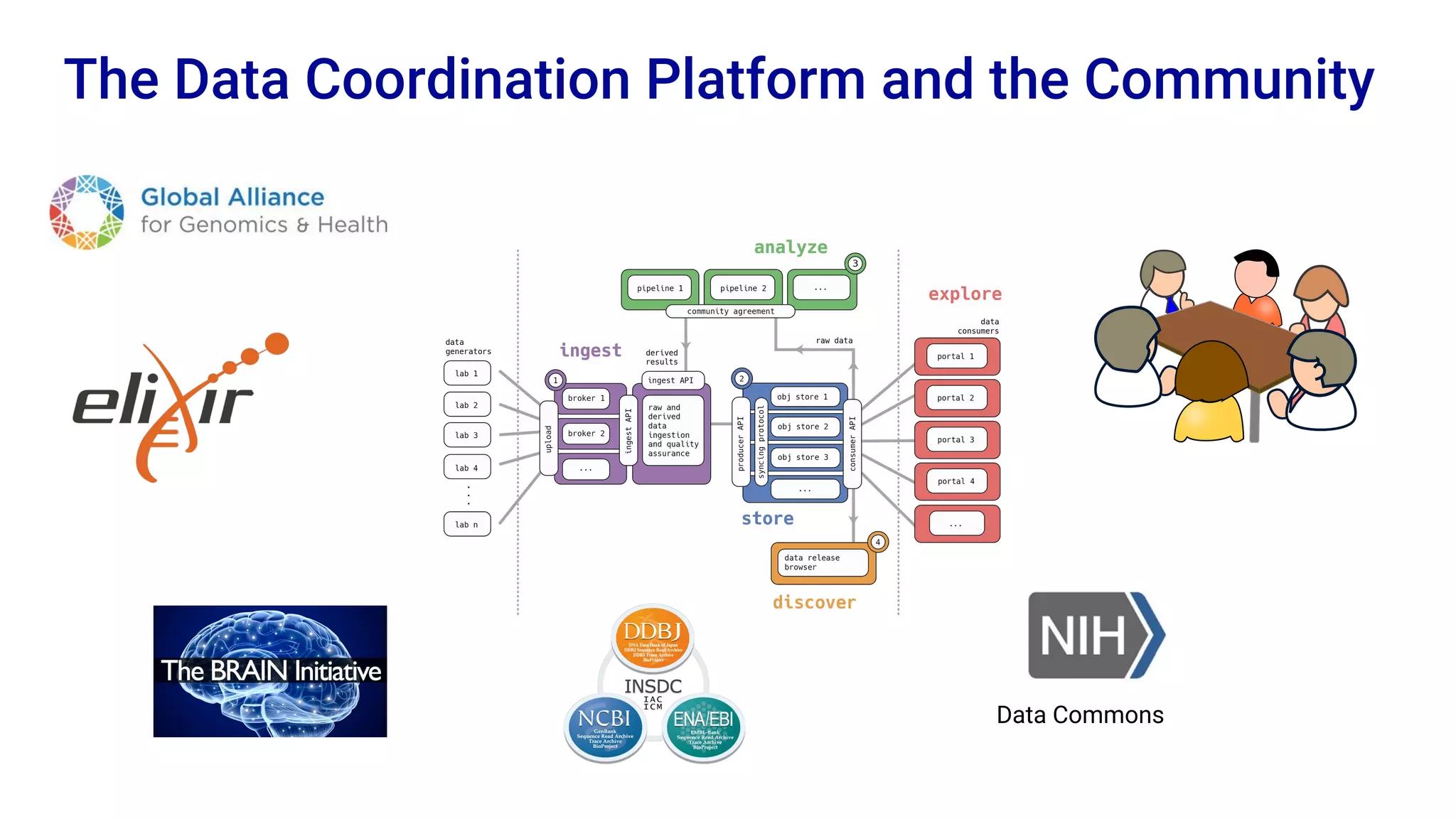
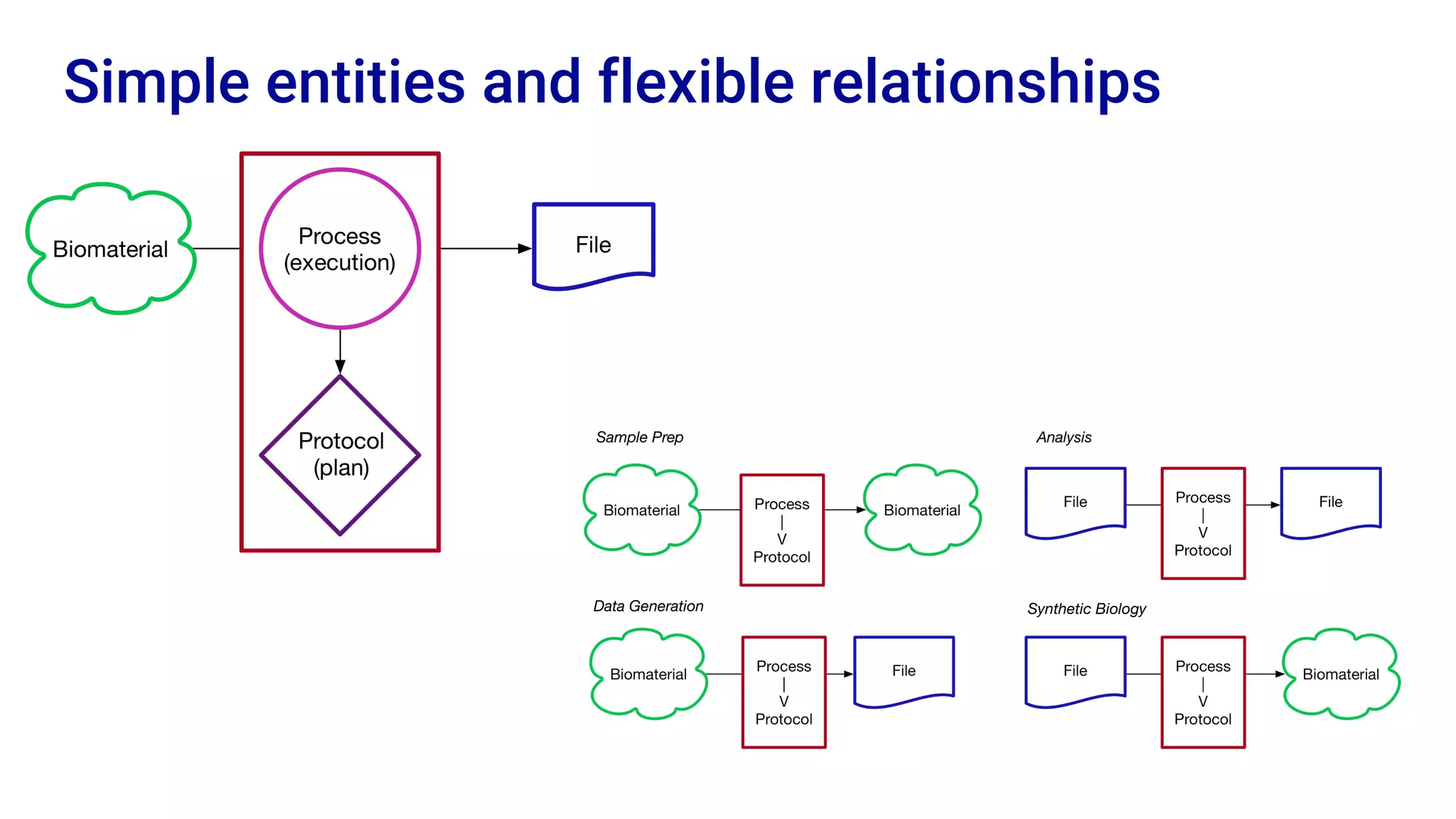
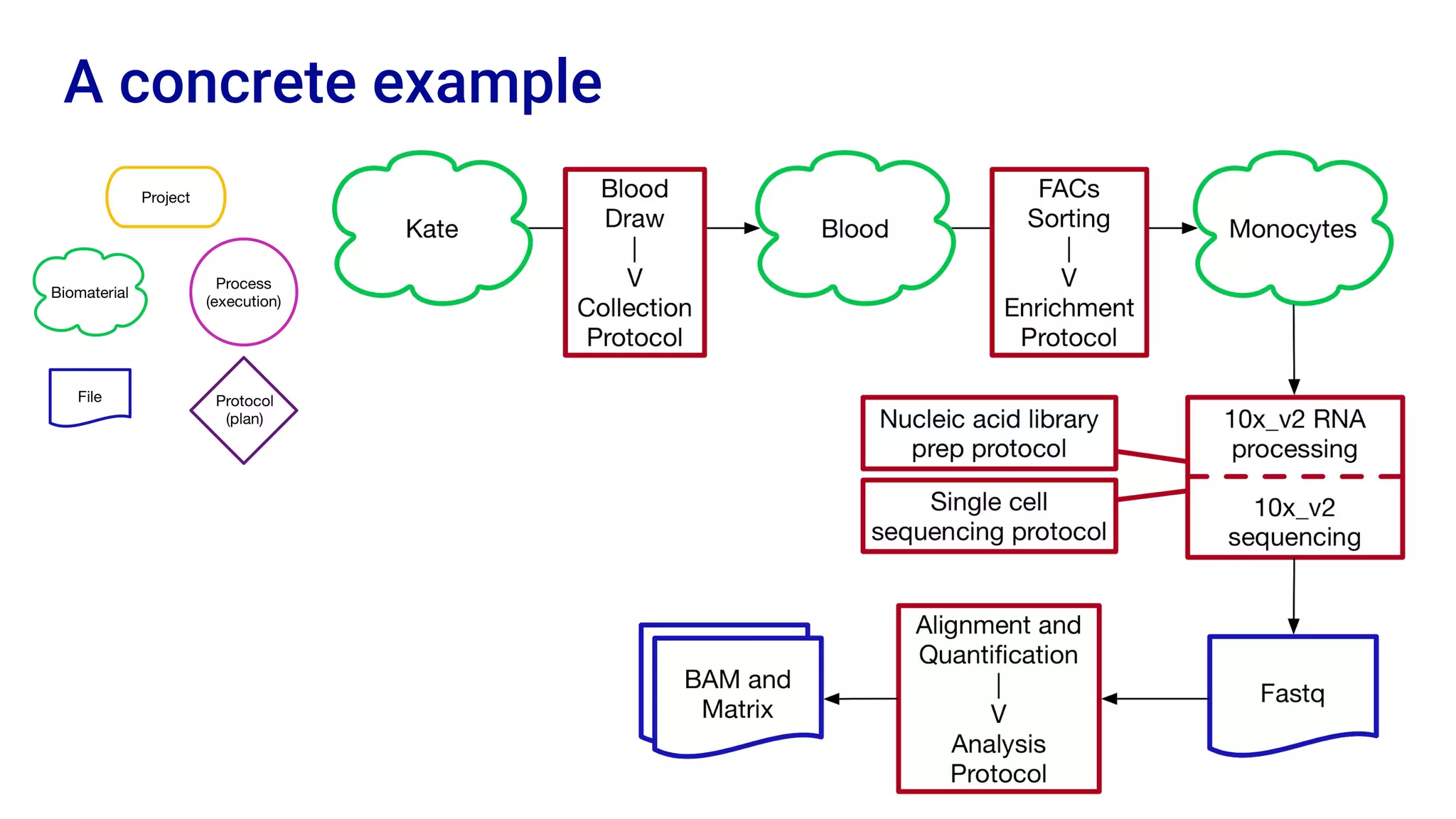
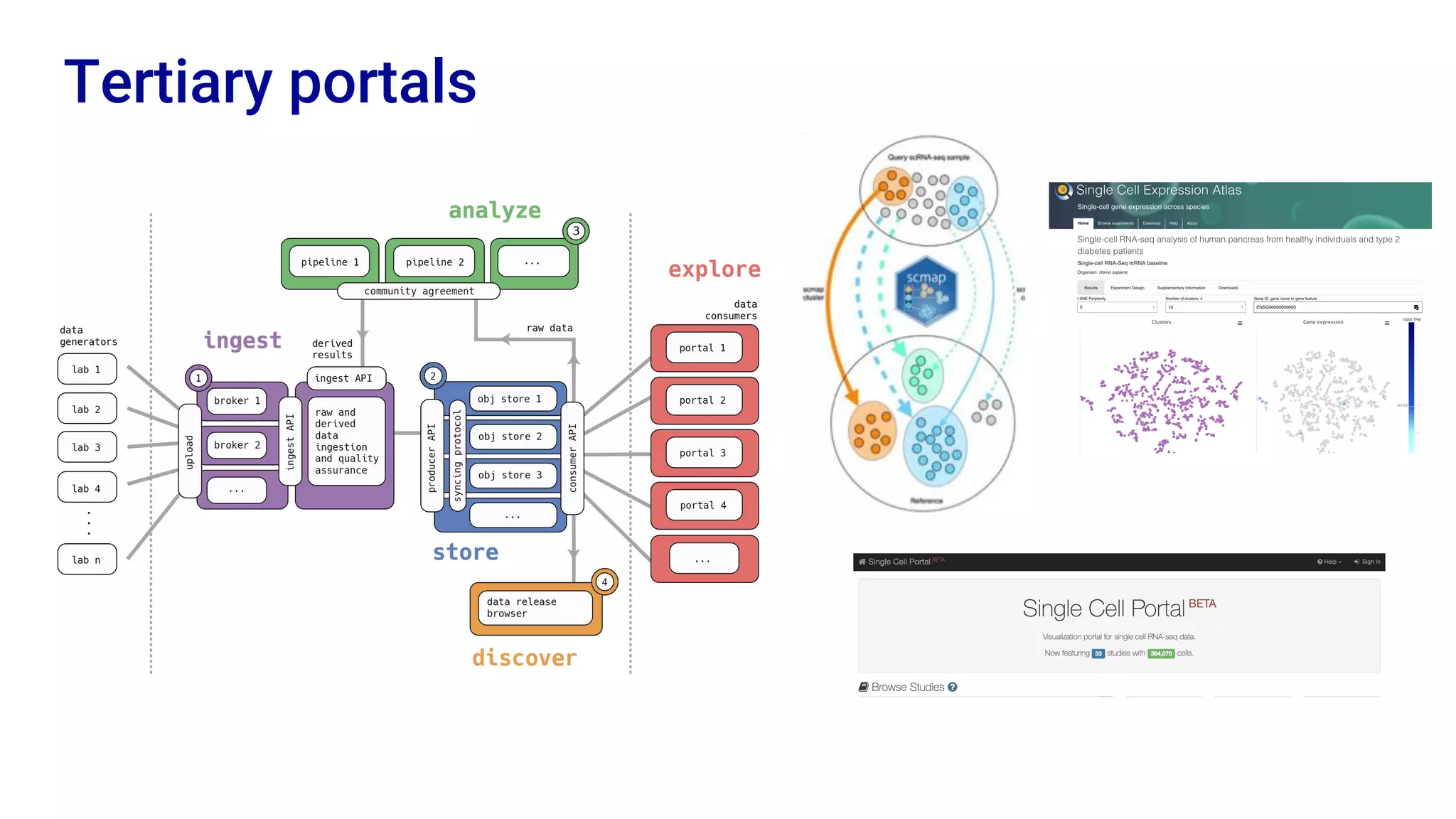
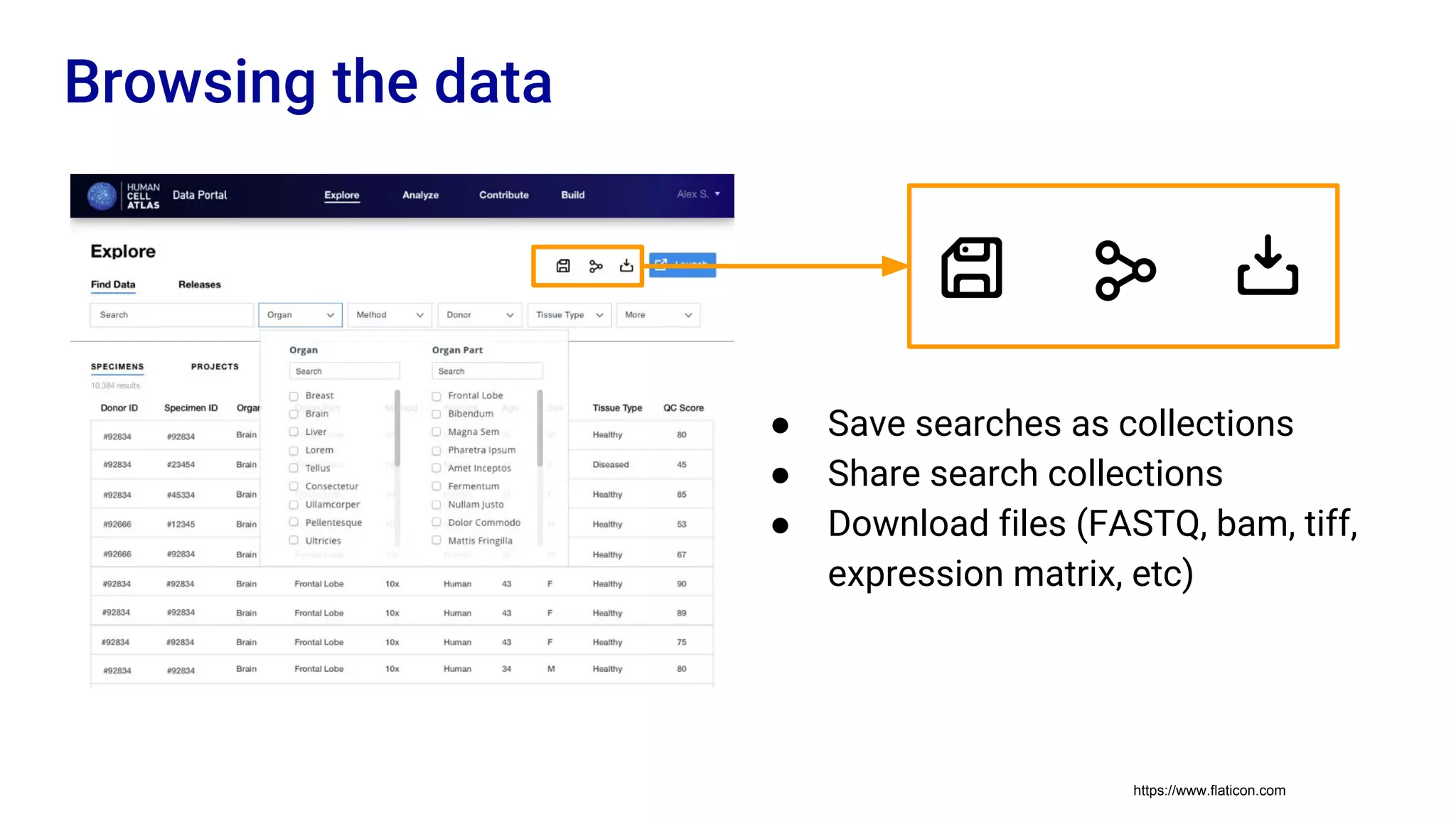
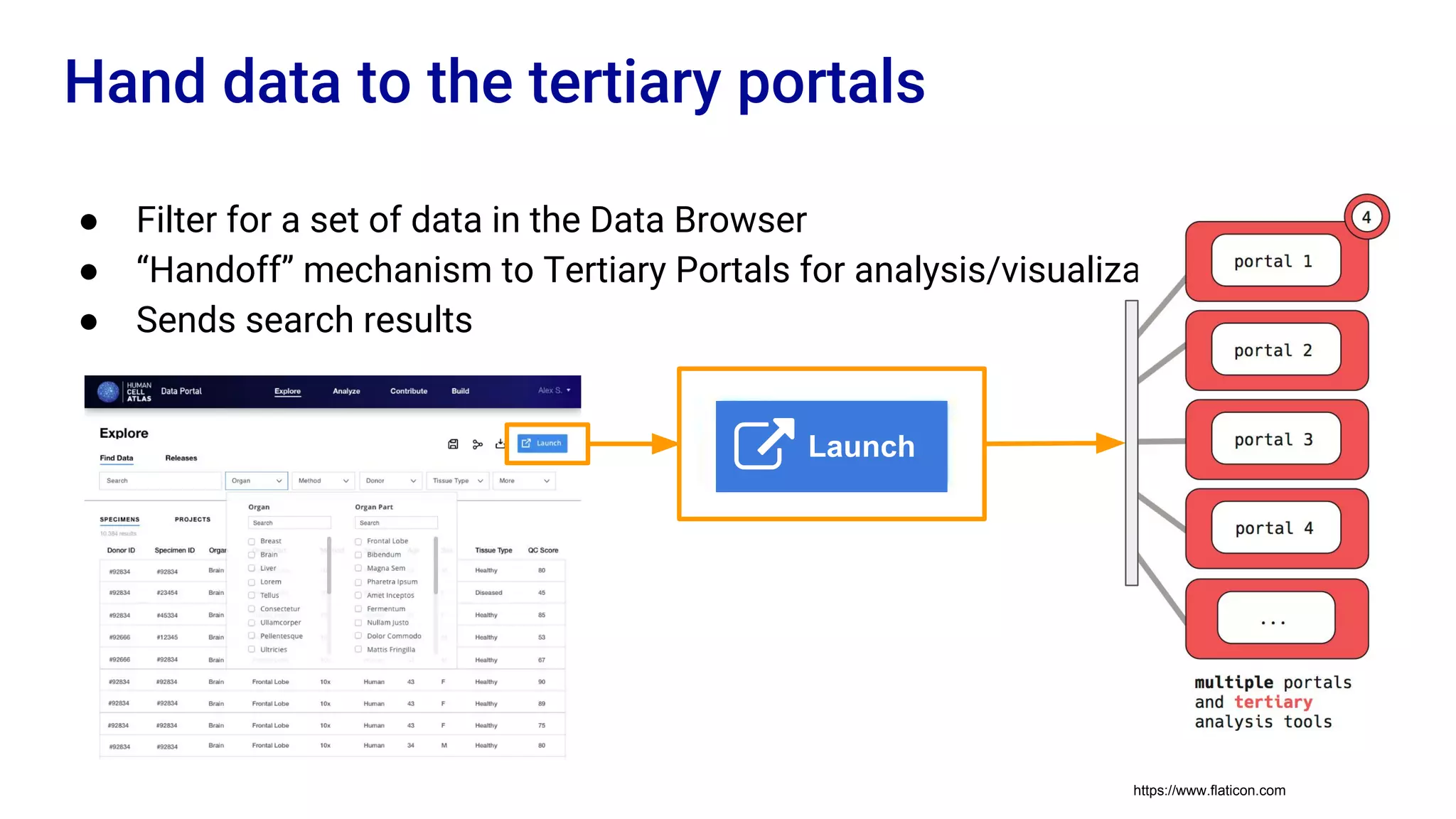
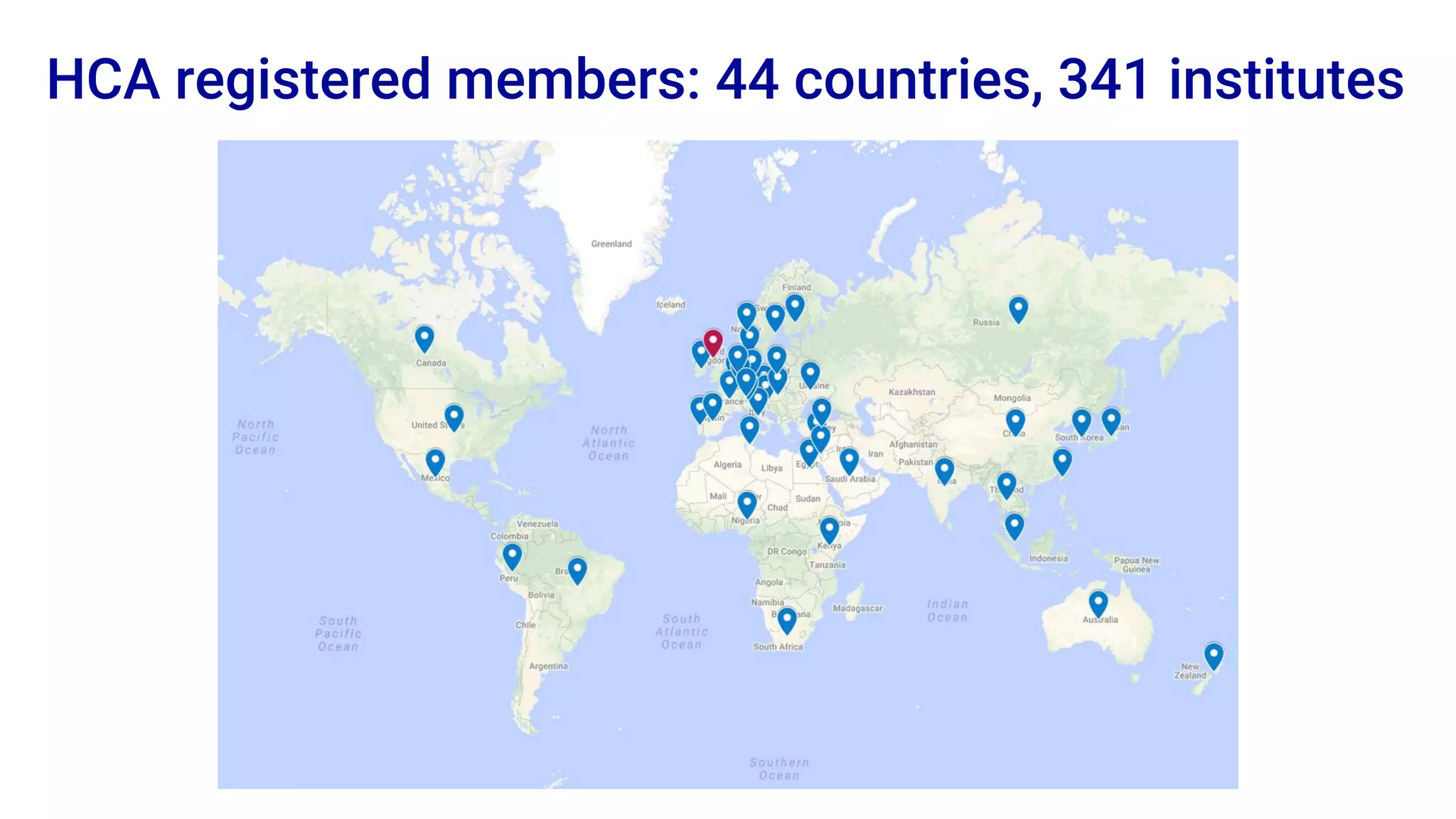
The Human Cell Atlas aims to create a comprehensive reference map of all human cell types and their properties, serving as a foundation for understanding health and disease. The Data Coordination Platform (DCP) facilitates the organization and accessibility of large data sets generated by multiple labs worldwide, ensuring that data is open and standardized. Future goals include further development of community-driven metadata standards and enhancing data accessibility for researchers through various tools and interfaces.