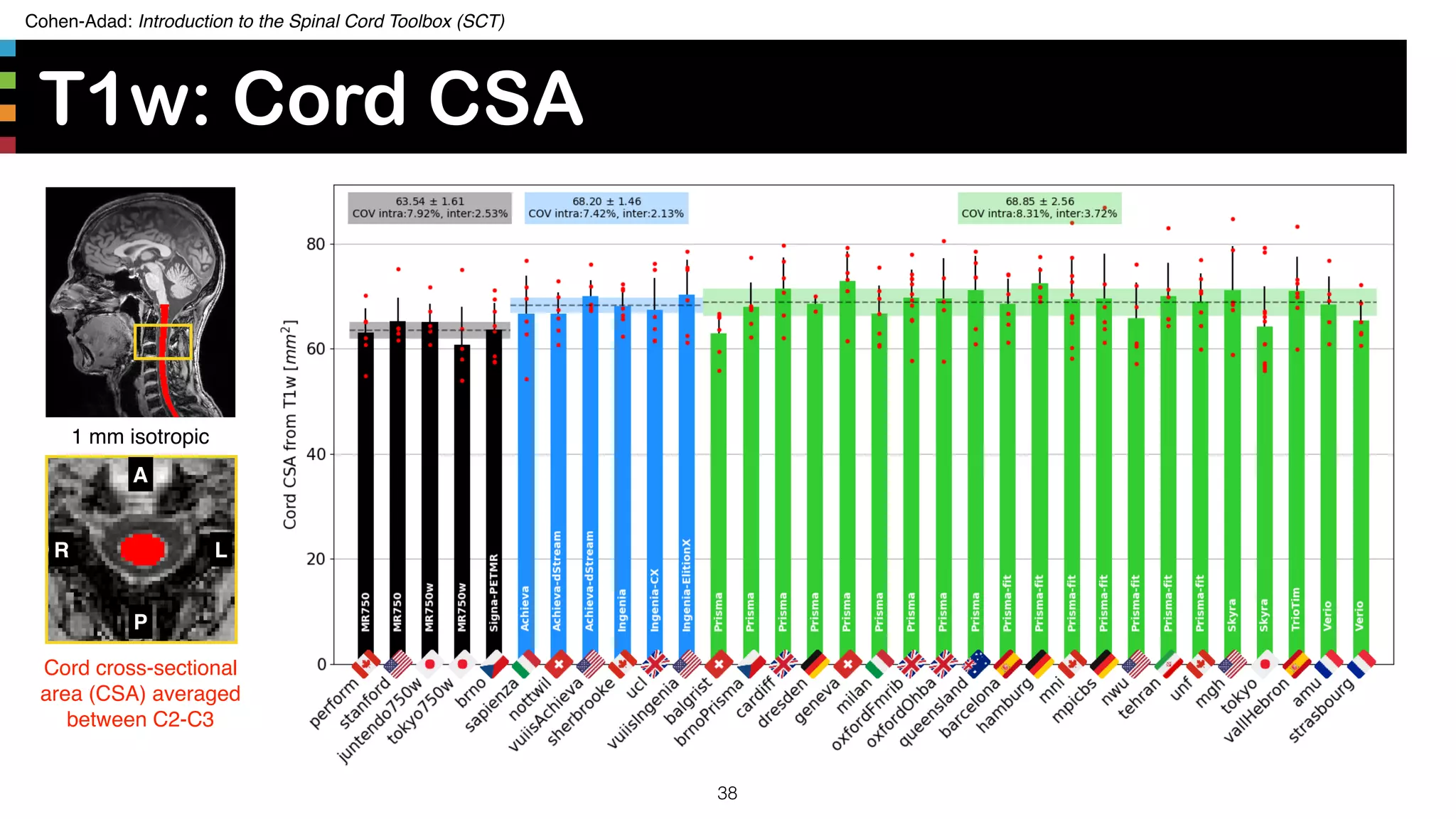
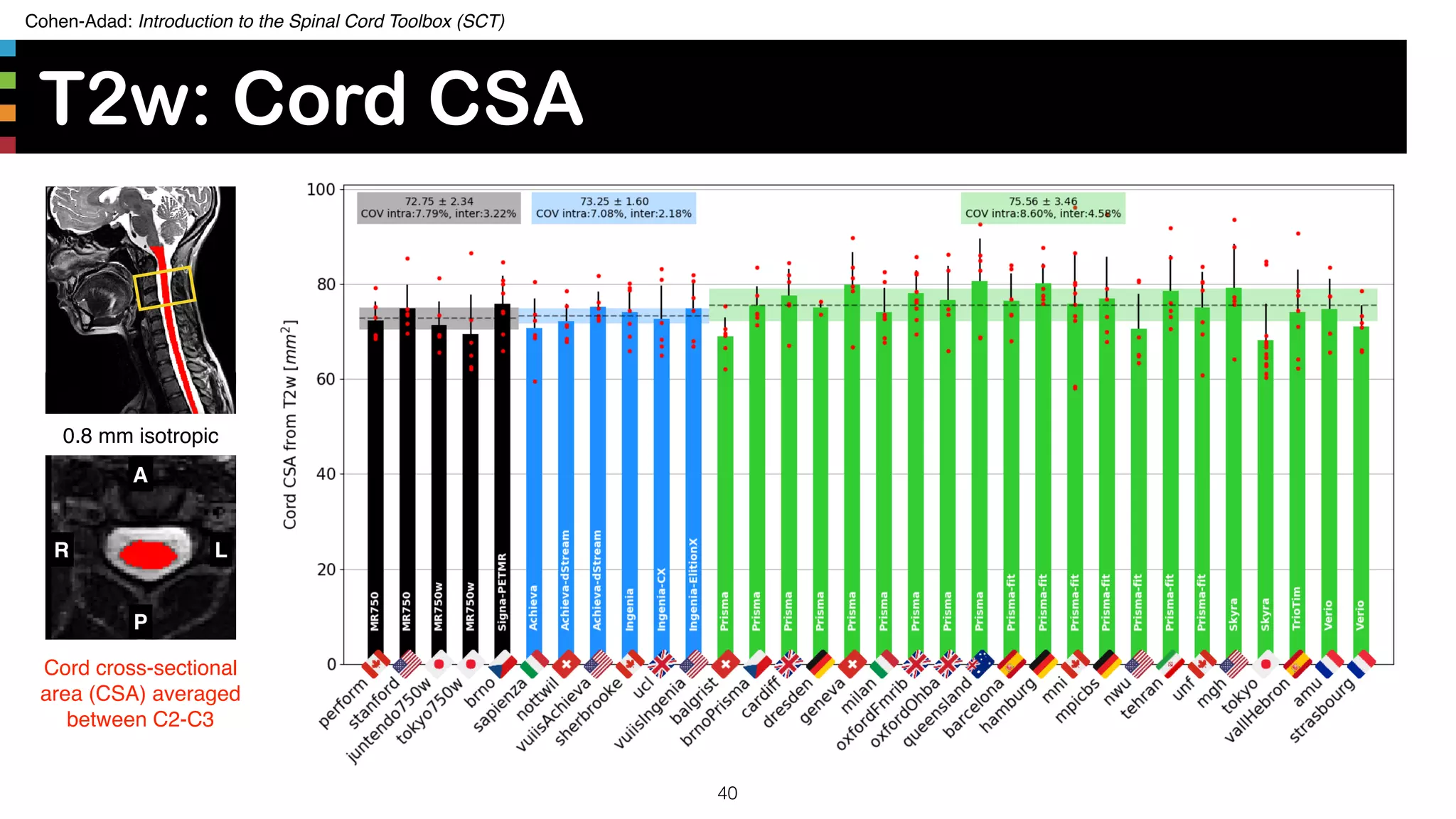
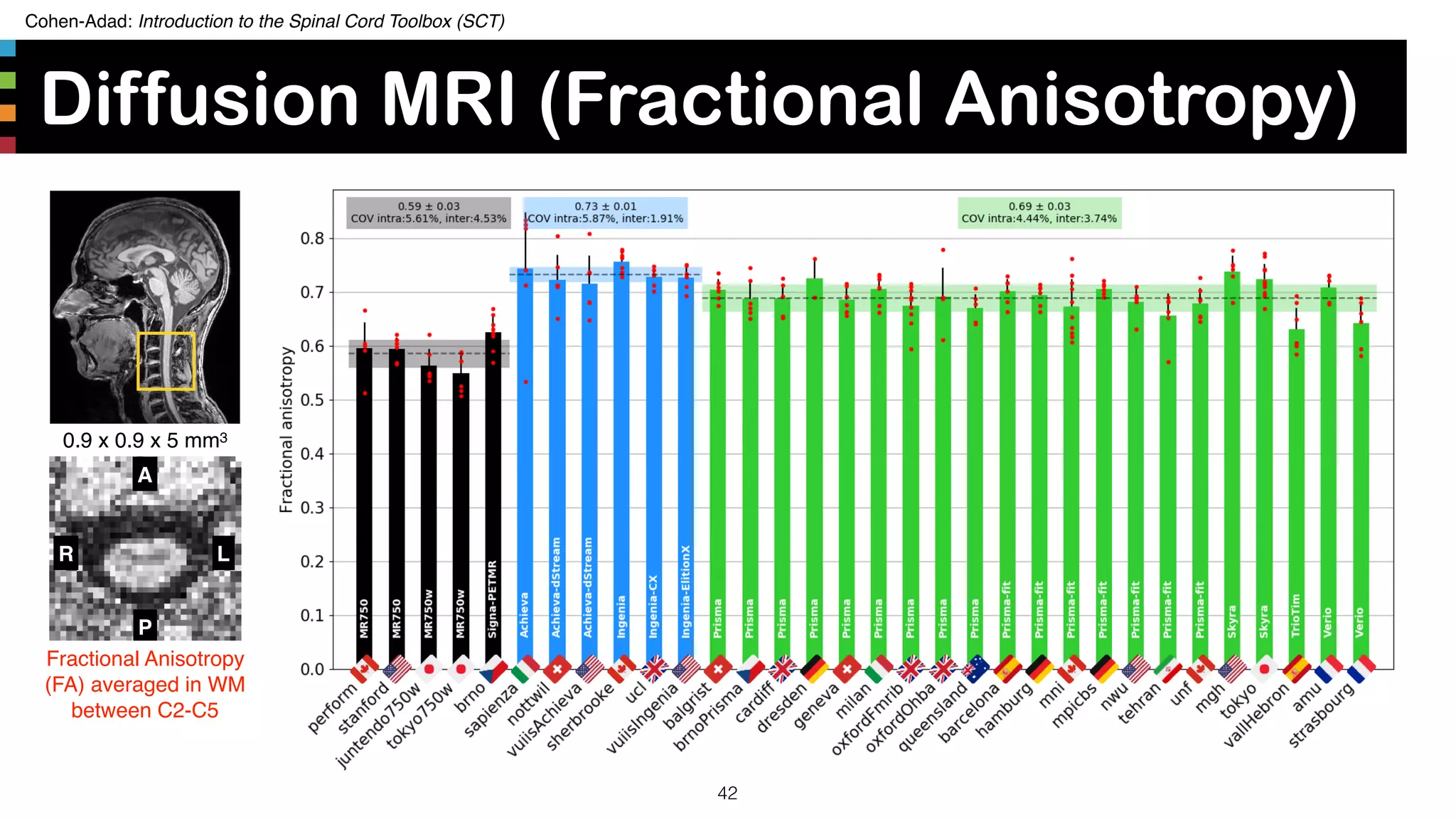
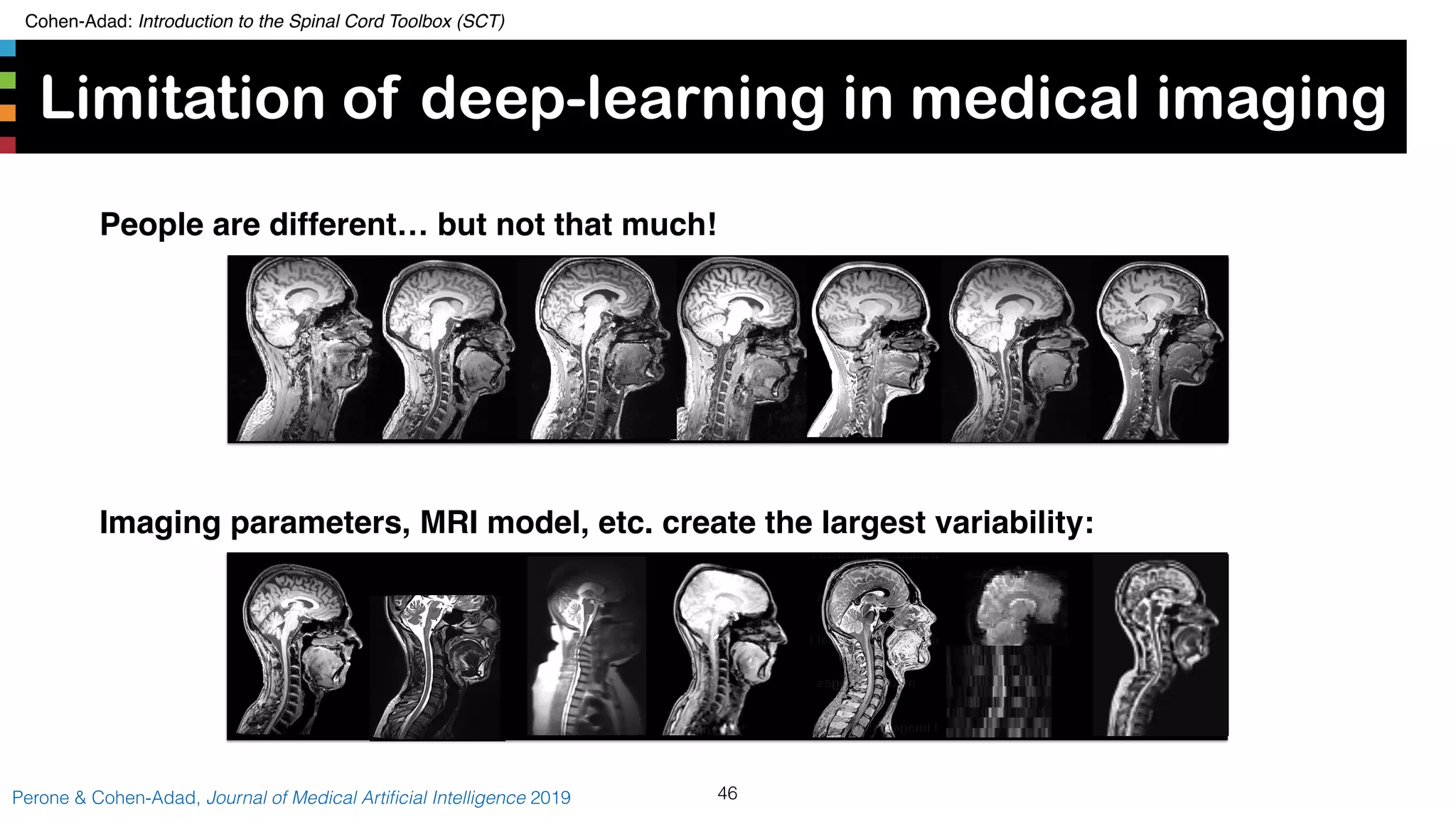
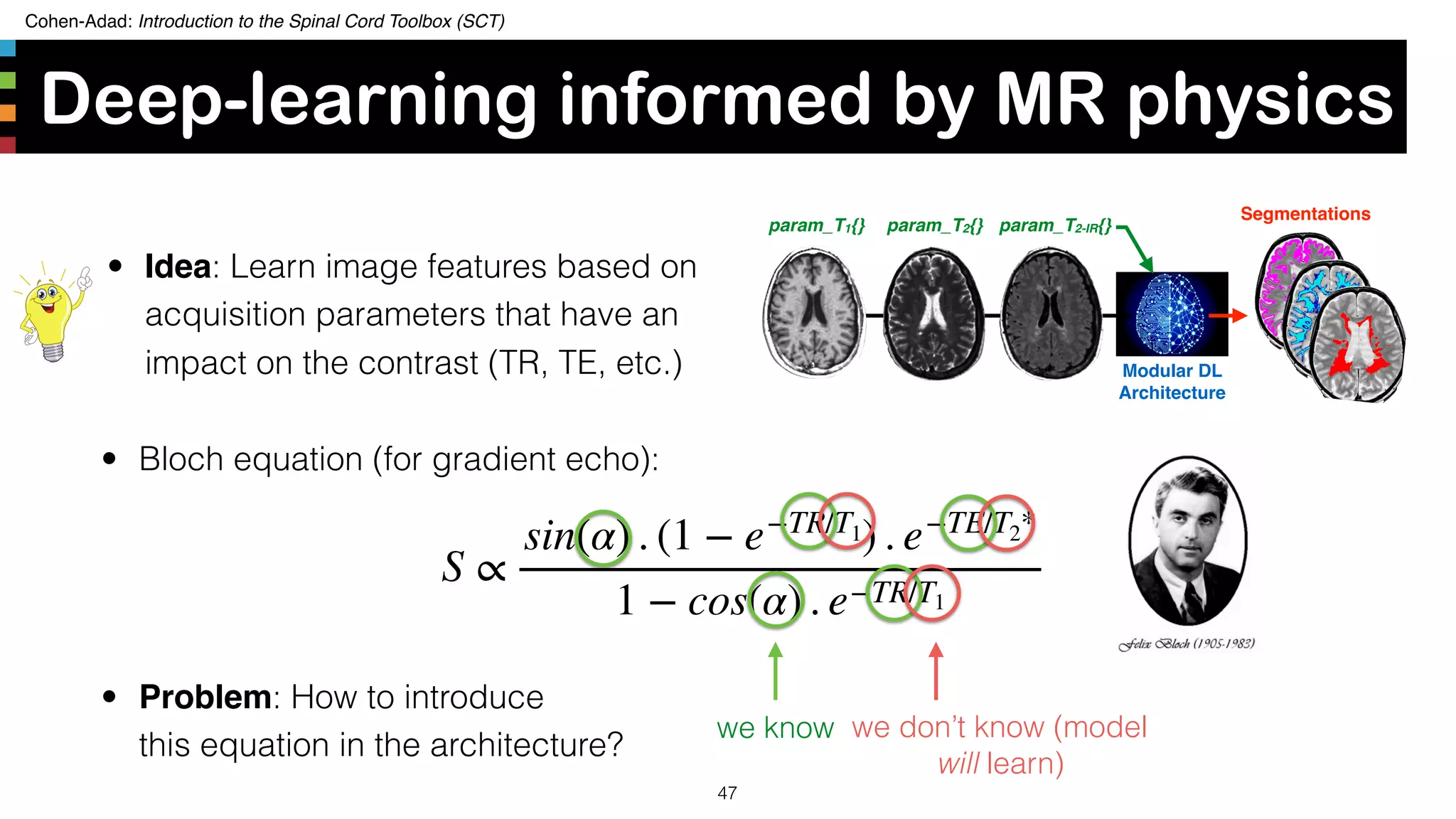
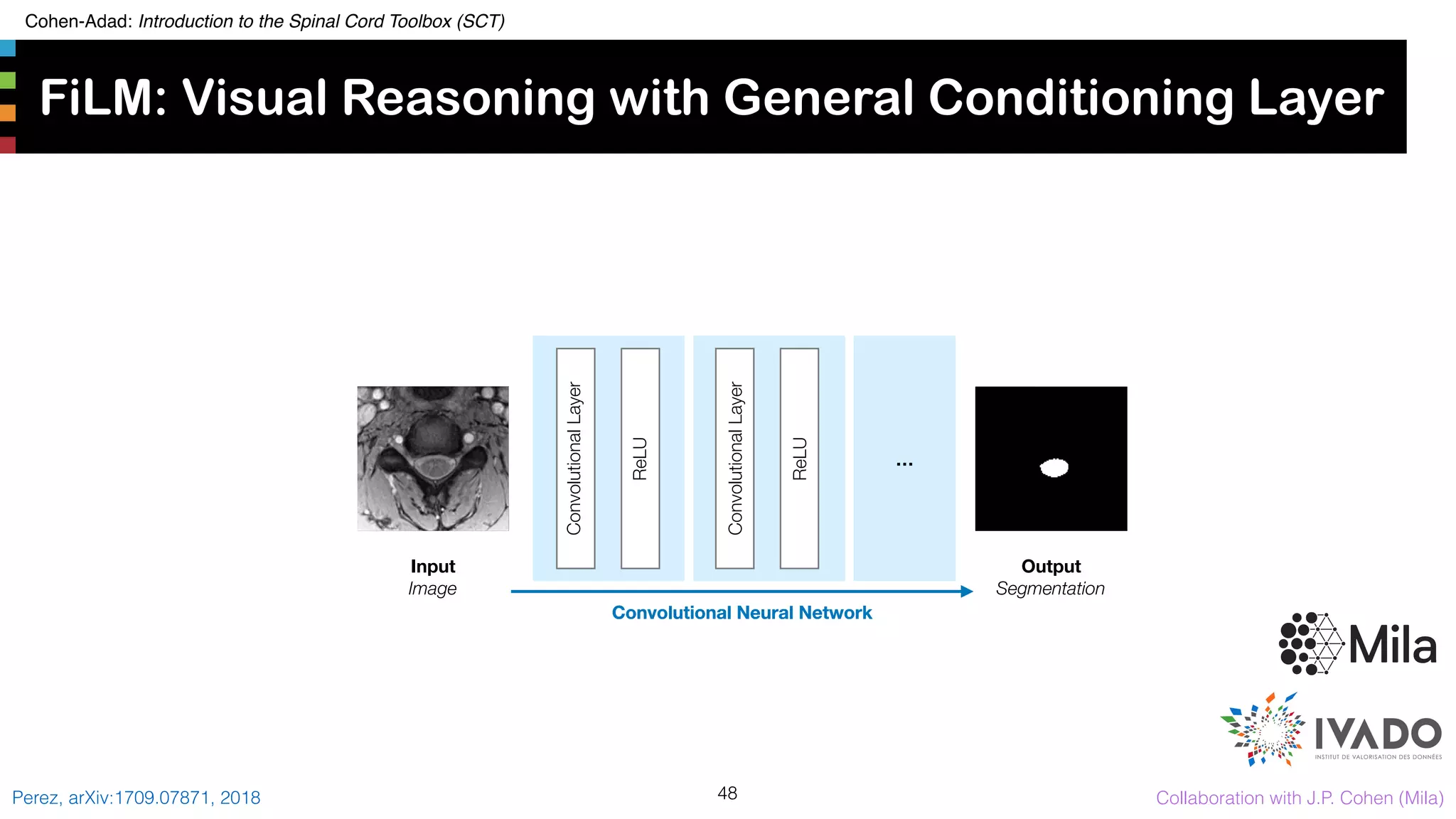
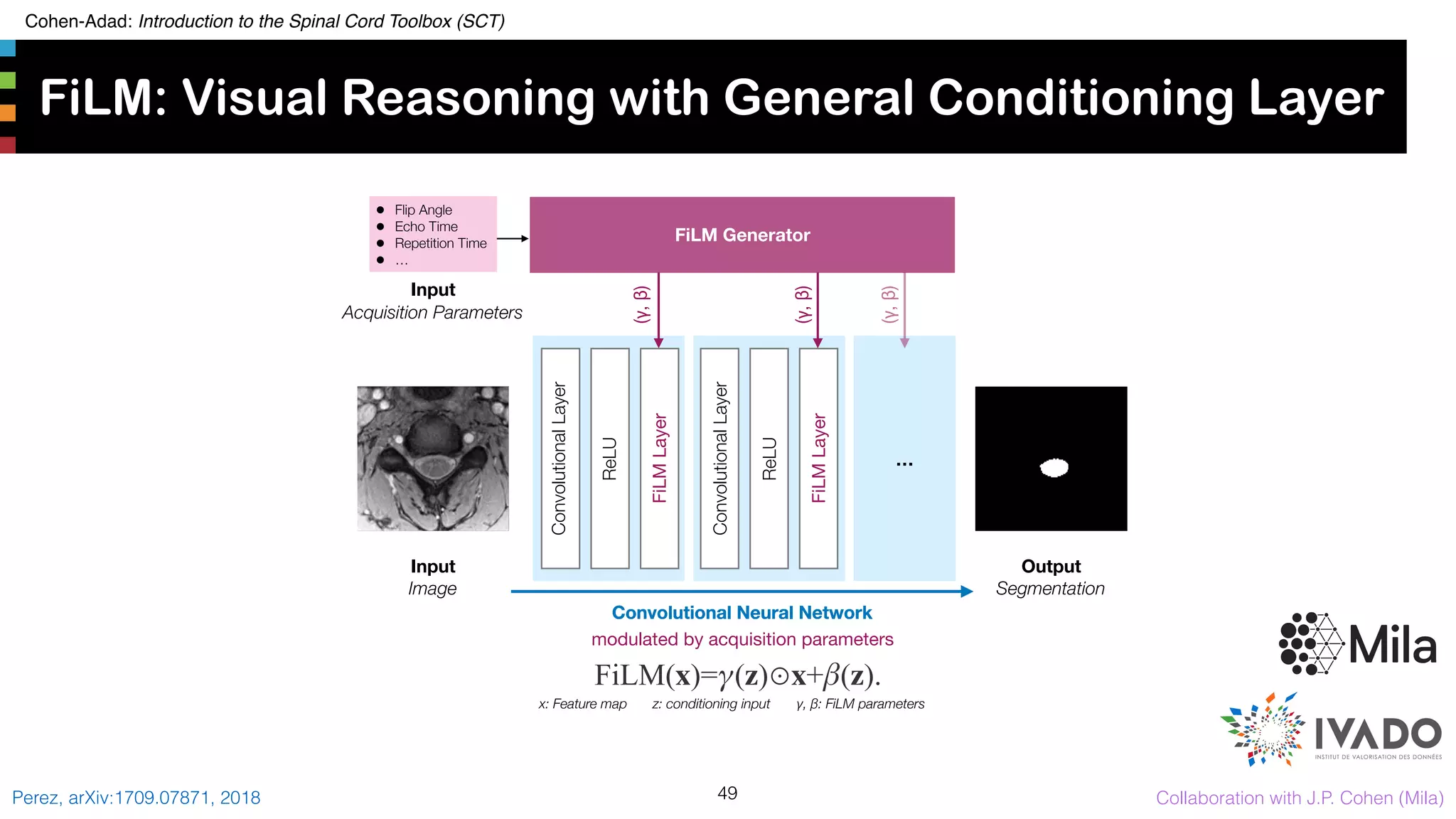
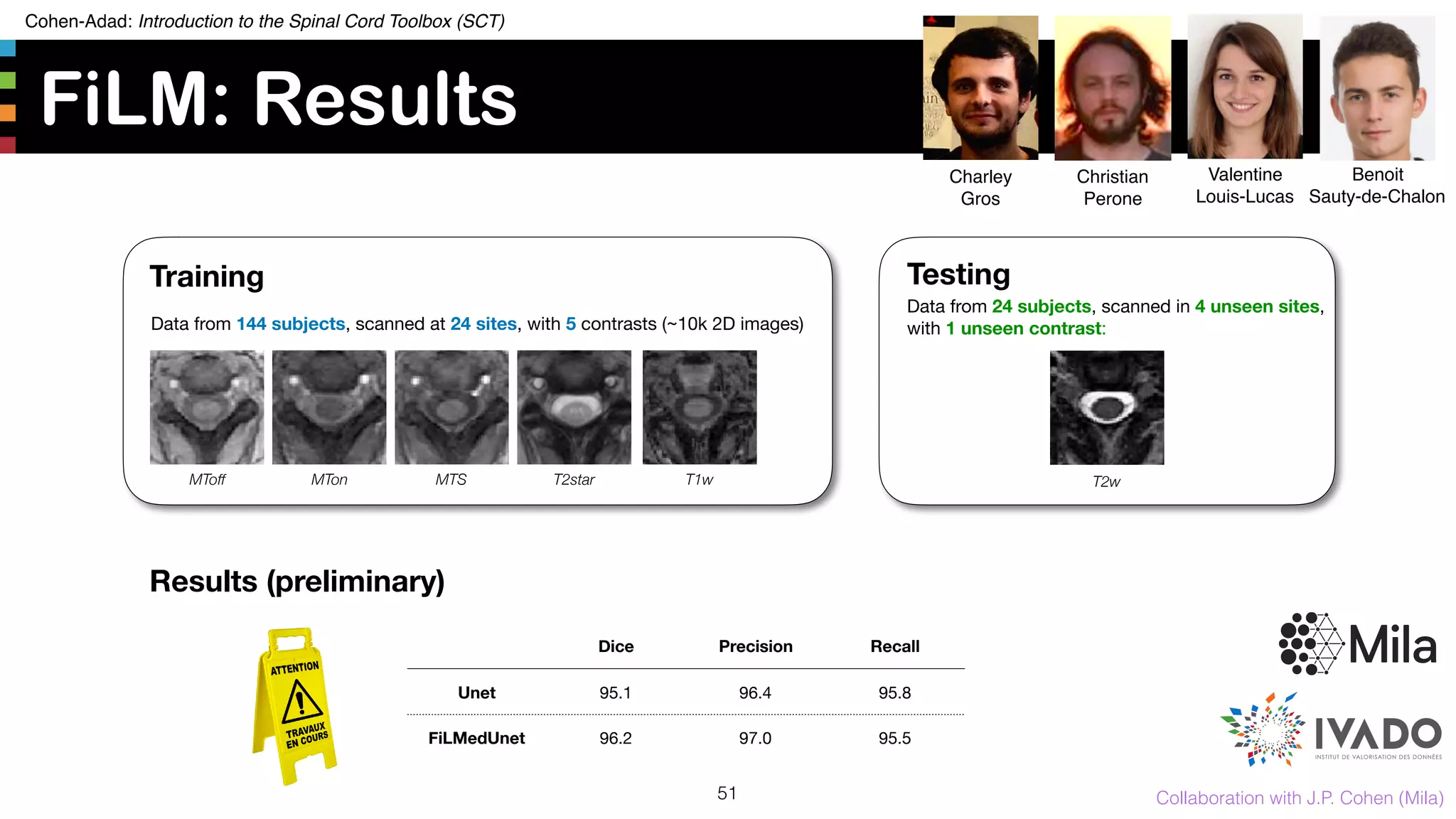
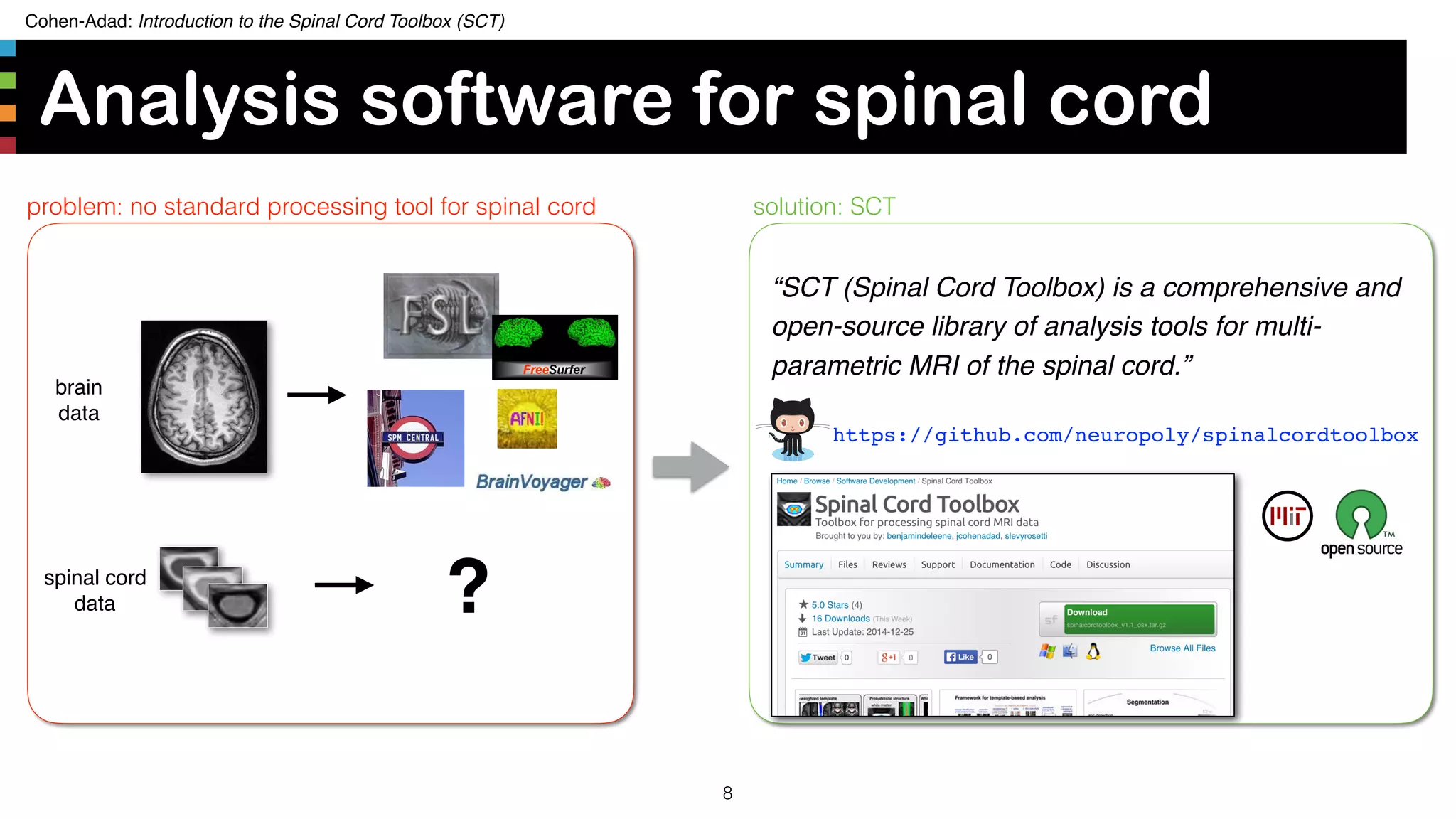
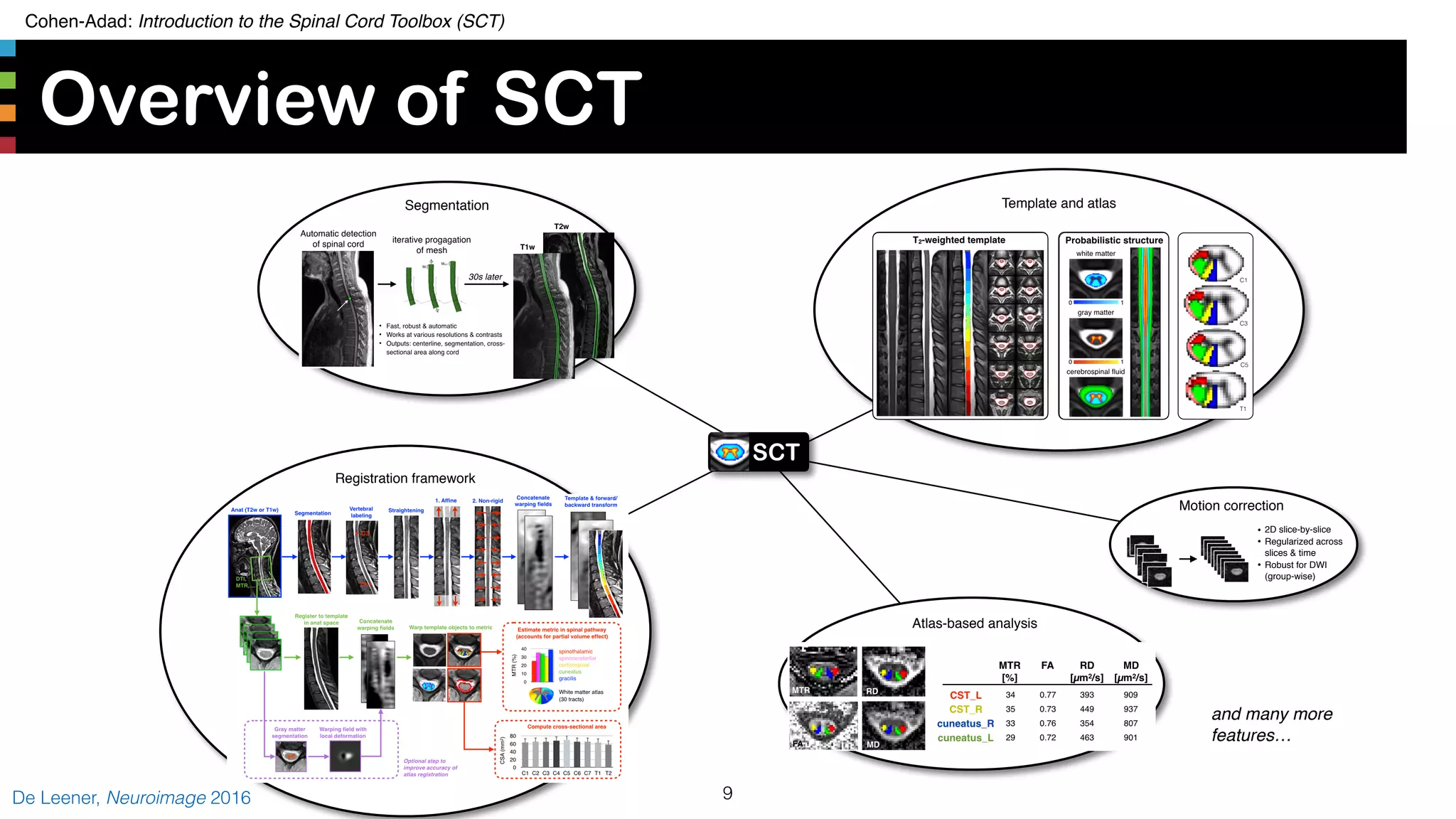
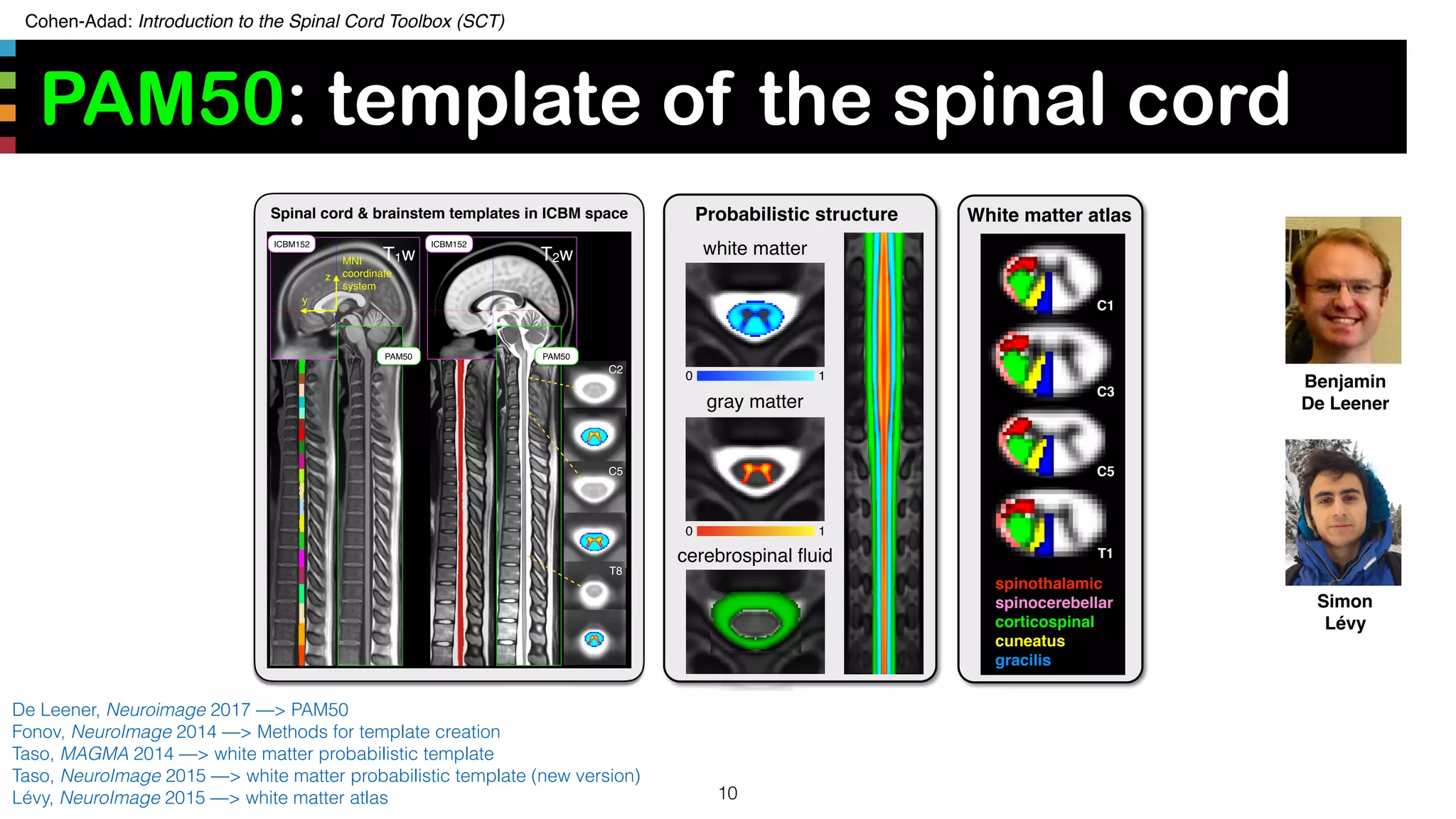
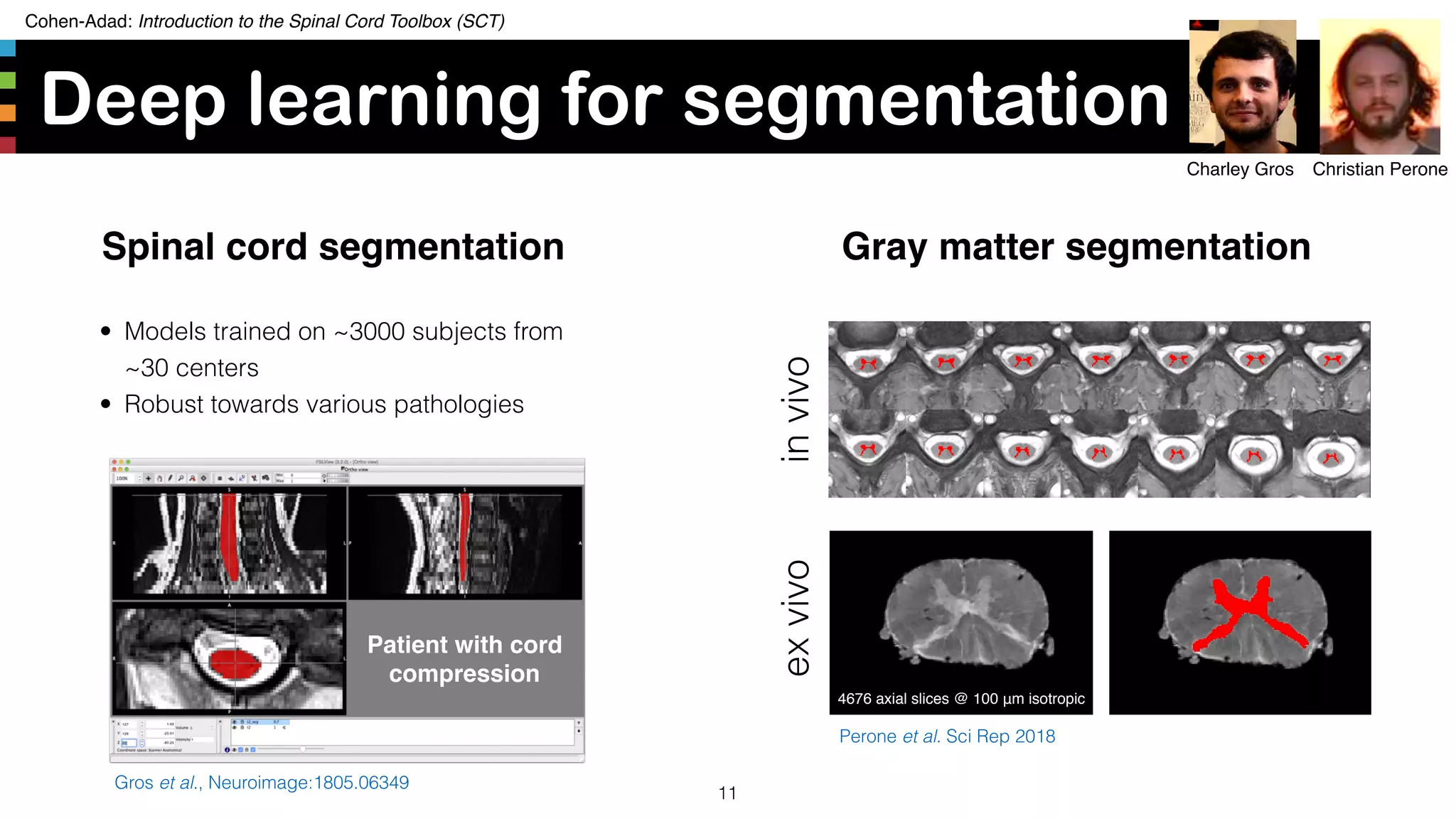
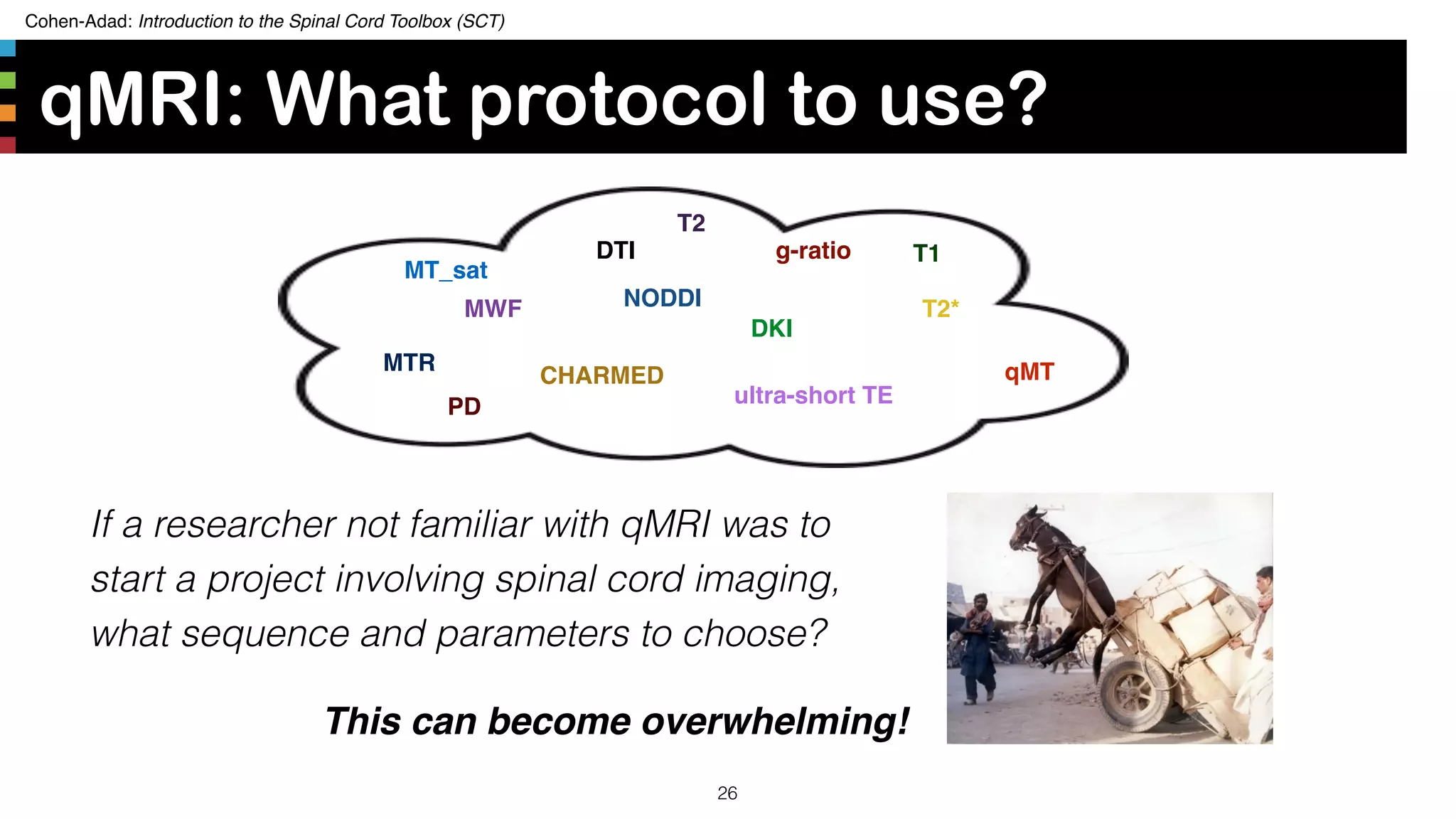
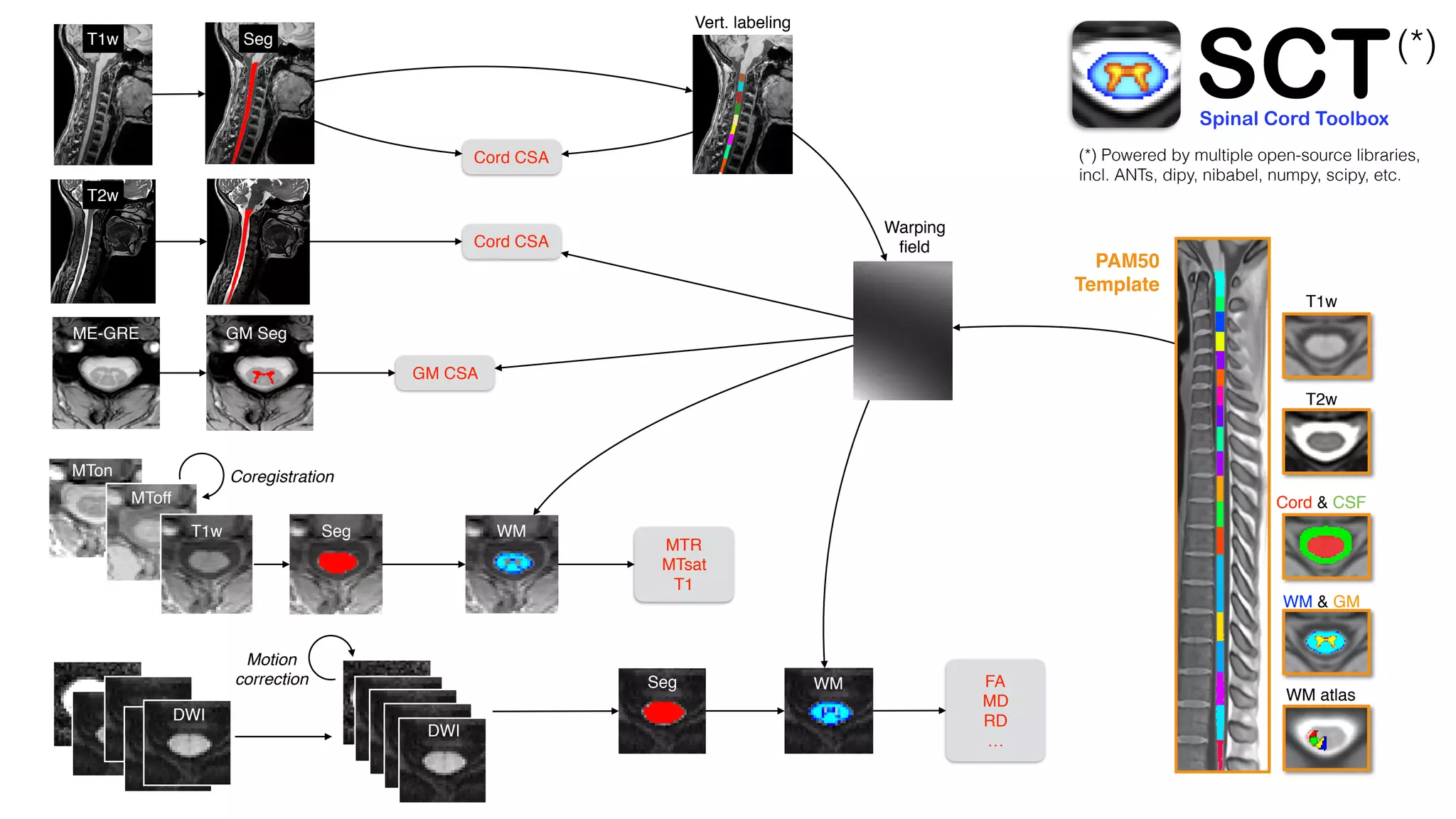
The document introduces the Spinal Cord Toolbox (SCT), an open-source software for analyzing quantitative MRI data of the spinal cord. SCT provides tools for segmenting the spinal cord and classifying tissue types, registering images to templates and atlases, and extracting quantitative metrics. It has been used in over 150 scientific publications for applications such as functional MRI of the spinal cord, studying microstructural changes with DTI/MT, and analyzing spinal cord shape/lesions in patients. The document also discusses efforts to standardize spinal cord MRI protocols through an initiative called the "Spine Generic Protocol" to facilitate multi-site studies.






![Cohen-Adad: Introduction to the Spinal Cord Toolbox (SCT)
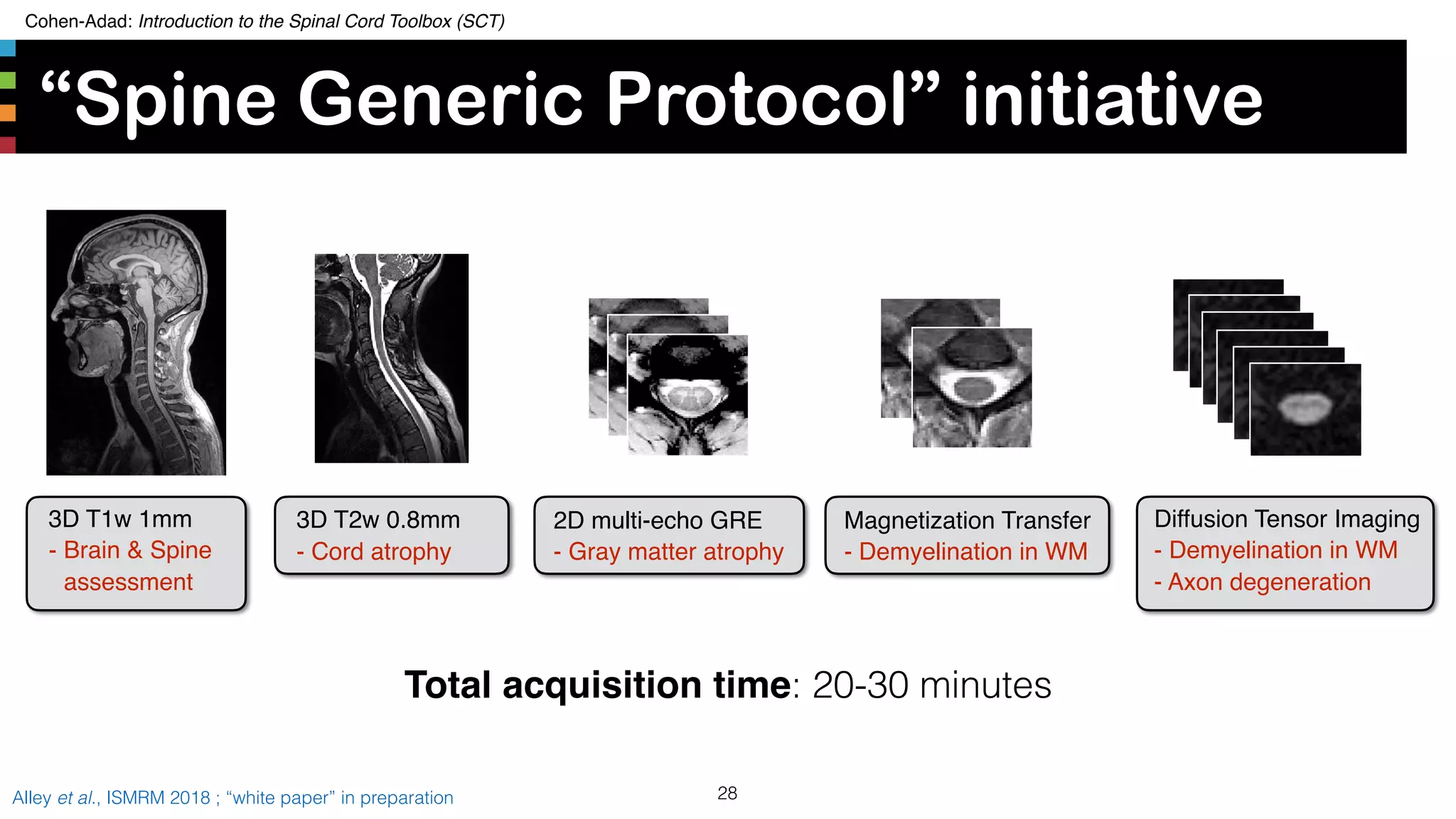
“Spine Generic Protocol” initiative
27Alley et al., ISMRM 2018 ; “white paper” in preparation
• Solution: Establish a consensus set of qMRI acquisition
parameters for the spinal cord at 3T. [Alley18]
• Similar to NINDS-CDE initiative, but focusing on qMRI
metrics and not specific to particular diseases
• 21 international sites involved in the optimization
• Protocol available for GE, Philips and Siemens at:
www.spinalcordmri.org/protocols](https://image.slidesharecdn.com/20190808sctintroduction-190810135751/75/SCT-course-27-2048.jpg)

![Cohen-Adad: Introduction to the Spinal Cord Toolbox (SCT)
Data structure: BIDS
31Gorgolewski et al. Sci Data 3, 160044 ; [2] https://github.com/bids-standard/bep001
http://bids.neuroimaging.io/ spineGeneric_multiSubjects
!"" dataset_description.json
!"" participants.json
!"" participants.tsv
!"" sub-ucl01
!"" sub-ucl02
!"" sub-ucl03
!"" sub-ucl04
!"" sub-ucl05
#"" sub-ucl06
!"" anat
$ !"" sub-ucl06_T1w.json
$ !"" sub-ucl06_T1w.nii.gz
$ !"" sub-ucl06_T2star.json
$ !"" sub-ucl06_T2star.nii.gz
$ !"" sub-ucl06_T2w.json
$ !"" sub-ucl06_T2w.nii.gz
$ !"" sub-ucl06_acq-MToff_MTS.json
$ !"" sub-ucl06_acq-MToff_MTS.nii.gz
$ !"" sub-ucl06_acq-MTon_MTS.json
$ !"" sub-ucl06_acq-MTon_MTS.nii.gz
$ !"" sub-ucl06_acq-T1w_MTS.json
$ #"" sub-ucl06_acq-T1w_MTS.nii.gz
#"" dwi
!"" sub-ucl06_dwi.bval
!"" sub-ucl06_dwi.bvec
!"" sub-ucl06_dwi.json
#"" sub-ucl06_dwi.nii.gz
What is BIDS?
BIDS is a convention for organizing neuroimaging
data files and folders
Why BIDS?
Easier to share data and to process them with
BIDS-compatible processing pipelines.](https://image.slidesharecdn.com/20190808sctintroduction-190810135751/75/SCT-course-31-2048.jpg)

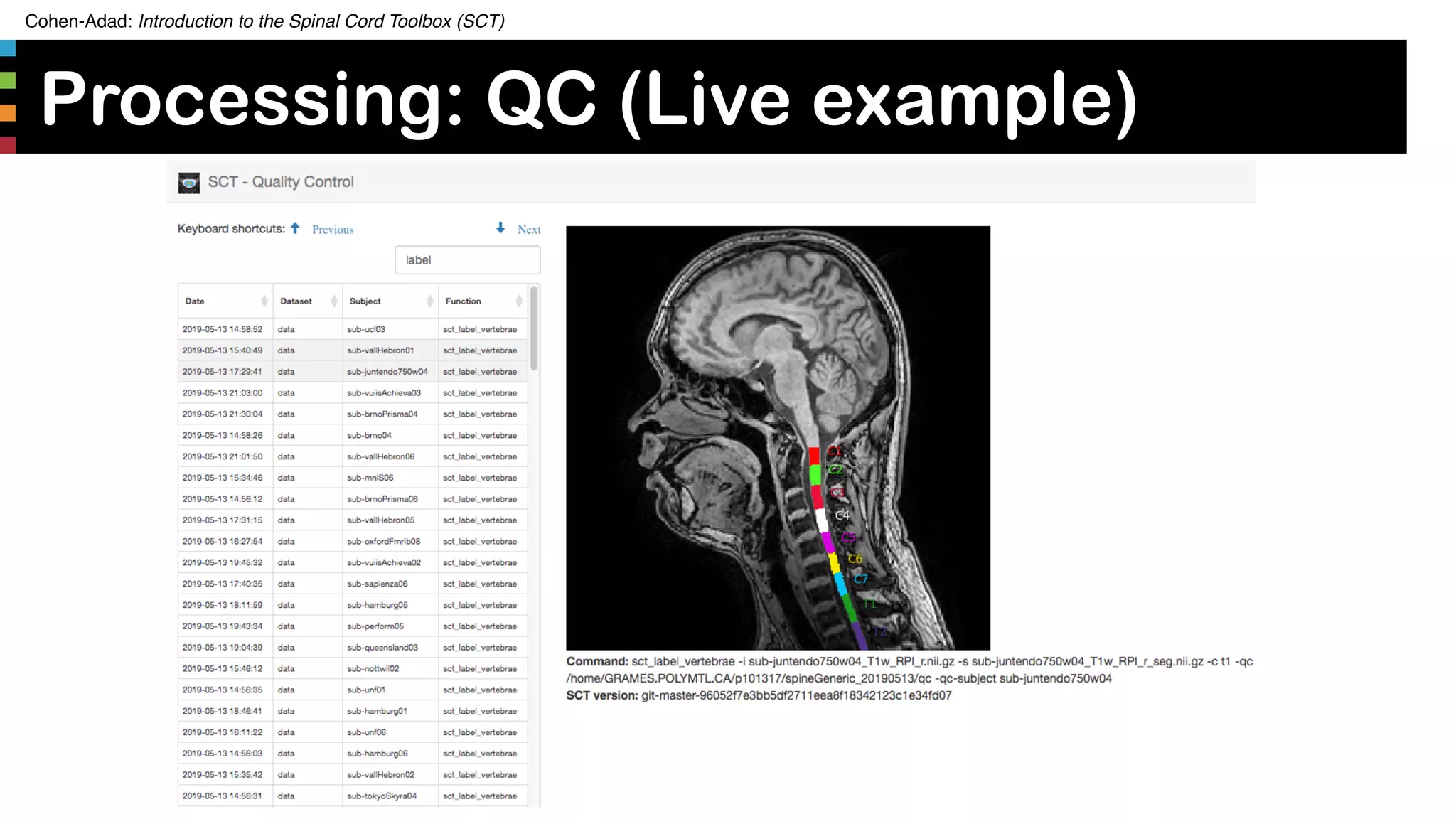
![Cohen-Adad: Introduction to the Spinal Cord Toolbox (SCT)
Manual correction
35https://github.com/sct-pipeline/spine-generic/blob/master/README.md#quality-control-rapid
• Number of subjects that needed
corrections:
• T1w seg: 17 (8%)
• T2w seg: 2 (1%)
• T2star GM seg: 15 (7%)
• T1w_ax: 8 (4%)
• Vertebral labeling (on T1w): 25 (12%)
• SOP for manual correction after
QC [1]](https://image.slidesharecdn.com/20190808sctintroduction-190810135751/75/SCT-course-35-2048.jpg)