RNA Nanostructures Design Characterization and Applications Methods in Molecular Biology 2709 Kirill A. Afonin (Editor)
RNA Nanostructures Design Characterization and Applications Methods in Molecular Biology 2709 Kirill A. Afonin (Editor)
RNA Nanostructures Design Characterization and Applications Methods in Molecular Biology 2709 Kirill A. Afonin (Editor)


















![will also discuss how MD simulations of two KL motifs can be used to
estimate the conformation and stability of RNA nanoring as well as to
explain the vibrational characteristics of RNA nanoring.
Key words Molecular dynamics simulations – RNA motif – RNA
nanostructure – AMBER – CHARMM – NAMD
1 Introduction
Since RNA tectoRNA [1–5] was built in the early 2000s, RNA
nanotechnology has rapidly developed computationally and
experimentally. The various shapes of RNA nanostructures have been
built using numerous RNA motifs. The examples of RNA motifs are kink-
turn motif, junction motif, pseudoknot, kissing loop hairpins, GNRA
loop-receptor, triple helical scaffold, and G- quadruplex. Examples of
RNA nanostructure shapes, which are constructed by RNA motifs, are
triangle [6–8], square [9], hexagonal ring [10–12], cubes [13–16], and
polyhedron [17, 18]. The various shapes of RNA nanostructures also
have been investigated to develop diverse biomedical applications, such
as drug delivery [19–21], gene therapy [14, 22], and molecular beacon
[23–31].
Several computational methods have been developed to design RNA
nanostructure. Examples of computational design tools are RNA2D3D
[32], NanoTiler [33], Assemble2 [34], INFO-RNA [35], and NUPACK
[36]. Once an initial RNA nanostructure is computationally generated,
molecular dynamics (MD) simulations can be used to fix steric crashes
in the nanostructure as well as to investigate the stability and
conformational changes of nanostructure. Most commonly used MD
simulation packages are Assisted Model Building with Energy
Refinement (AMBER, https://
ambermd.
org/
) [37], Nanoscale
Molecular Dynamics (NAMD, https://
www.
ks.
uiuc.
edu/
) [38],
Chemistry at Harvard Macromolecular Mechanics (CHARMM, https://
www.
charmm.
org/
) [39], and GROningen MAchine for Chemical
Simulations (GROMACS, https://
www.
gromacs.
org/
) [40]. Each MD
simulation package uses specific atomic interaction parameters and
topological information, which are called force fields (FF). AMBER,
CHARMM, and GROMACS have their own FF, and the NAMD platform](https://image.slidesharecdn.com/111375-250318012721-6c51170a/75/RNA-Nanostructures-Design-Characterization-and-Applications-Methods-in-Molecular-Biology-2709-Kirill-A-Afonin-Editor-20-2048.jpg)

![both AMBER and CHARMM FFs. In Subheading 3.1, it will be explained
how to apply AMBER FF to prepare the initial solvated system and
topology file for a linear RNA motif. In Subheading 3.2, it will be
explained how CHARMM FF can be applied to a bent RNA motif and an
RNA nanostructure. The MD simulations of these systems will be
performed by NAMD package. For post MD simulation data analysis, we
will introduce the use of AMBER cpptraj program [41] and visual
molecular dynamics (VMD) [42]. The cpptraj is a very convenient and
powerful text command for data analysis, while VMD provides basic
data analysis tools based on a graphic interface. The detailed
explanations of cpptraj and VMD are described in Subheading 3.3.
In Note 1, we will introduce how MD simulation can be failed due to
the small periodic boundary box, especially when the biomolecule
experiences a large conformational change during MD simulations. In
Notes 2 and 3, we will also briefly describe how MD simulations of RNA
motifs can be used to estimate the conformational stability of RNA
nanostructure as well as to understand its vibrational mode.
2 Materials
In this section, we briefly introduce AMBER, CHARMM, and NAMD. We
will also briefly introduce VMD and Discovery Studio Visualizer (DSV),
which can be used for the visualization of molecular structure and MD
trajectory, molecular editing, and basic data analysis.
2.1 AMBER
AMBER (https://
ambermd.
org/
) is a comprehensive biomolecular MD
simulation package. The most recent AMBER FFs can be found in the
AMBER website (https://
ambermd.
org/
AmberModels.
php). However,
it is strongly advised that users check the details of FF in the most
recent AMBER reference manual (https://
ambermd.
org/
Manuals.
php)
before preparing MD simulation. AMBER MD simulation can be
performed by one of two commands, sander (simulated annealing with
NMR-derived energy restraints) or pmemd (particle mesh Ewald
molecular dynamics). The pmemd command shows better performance
in terms of computation speed. In addition, the performance of MD
simulation can be further improved when the graphics processing unit](https://image.slidesharecdn.com/111375-250318012721-6c51170a/75/RNA-Nanostructures-Design-Characterization-and-Applications-Methods-in-Molecular-Biology-2709-Kirill-A-Afonin-Editor-22-2048.jpg)


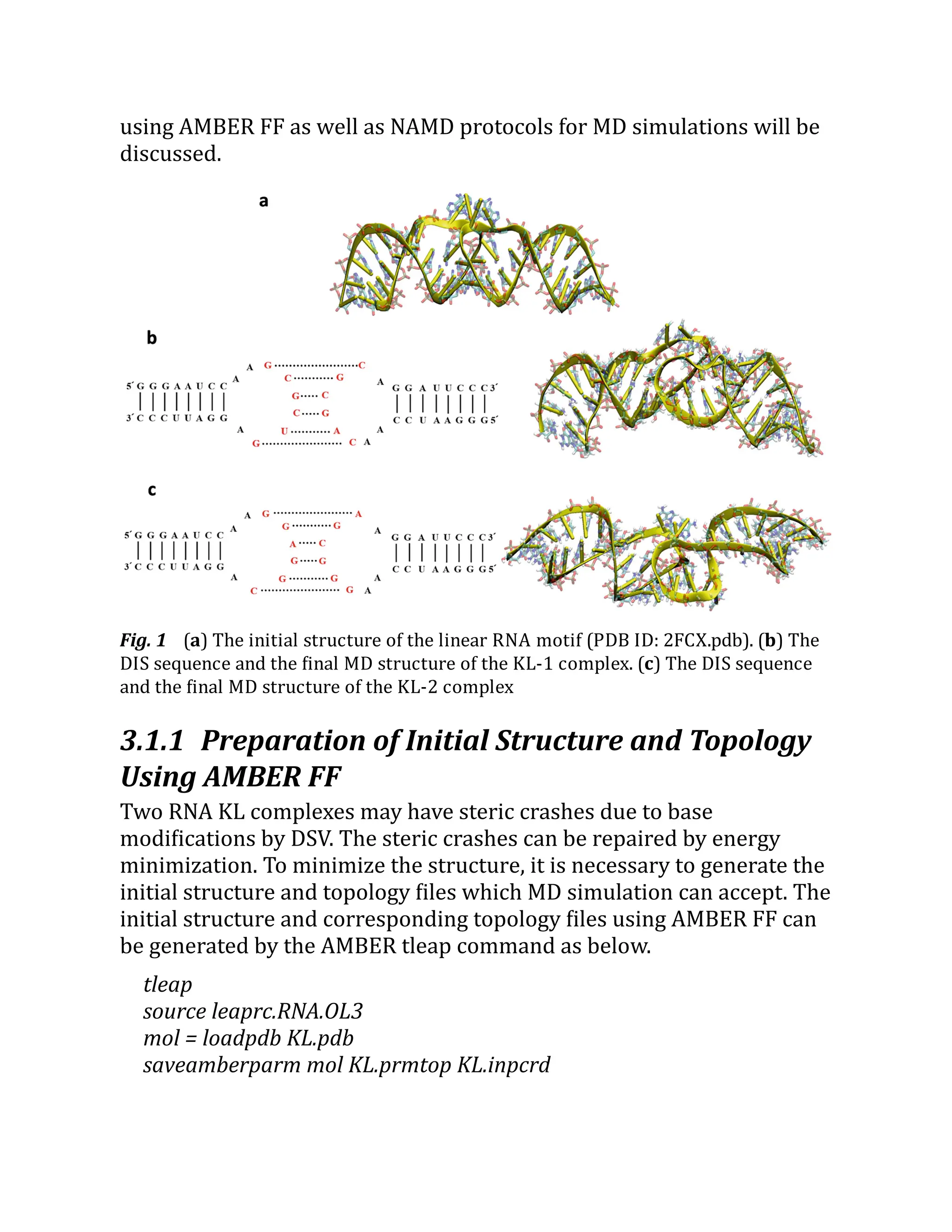


![identifies fixed atoms. To generate the pdb file for fixed atoms, in VMD,
select File → New Molecule → Browse → select KL-Solvated.prmtop →
select AMBER7 Parm → Browse → select KL-Solvated.inpcrd → select
AMBER7 Restart → Load. Then, type below commands to VMD console
window.
vmd > set all [atomselect top all]
vmd > set Fixatom [atomselect top "resname A C G U G5 C3"]
vmd > $all set beta 0
vmd > $Fixatom set beta 1
vmd > $all writepdb KL-Fixed.pdb
vmd > set center [measure center $all]
The above commands assign index 1 to the beta column of atoms
that belong to the residue names, A, C, G, U, G5, and C3. Atoms with
index 1 in the beta column are recognized as fixed atoms during NAMD
simulations. The result is saved to pdb format (KL-Fixed.pdb). The
command set center detects the center of the water box in (x, y, z)
coordinates. This coordinate will be used for the periodic boundary
conditions in the NAMD config file (line 48 in the EminEQ-I.conf in Table
1).
Table 1 The list of input files for minimization, equilibration, and MD simulations.
These files can be used to run NAMD simulations with AMBER FF. If the system is
prepared by CHARMM FF, use the config files listed in Table 2](https://image.slidesharecdn.com/111375-250318012721-6c51170a/75/RNA-Nanostructures-Design-Characterization-and-Applications-Methods-in-Molecular-Biology-2709-Kirill-A-Afonin-Editor-28-2048.jpg)




![KL-EminEQ-I.coor, KL-EminEQ-I.vel, KL-EminEQ-I.xst, KL-EminEQ-
I.restart.xsc.
KL-EminEQ-I.restart.coor, KL-EminEQ-I.restart.vel, KL-EminEQ-
I.restart.xsc, and KL-EminEQ-I.dcd.
The first group of files are the final coordinates, velocities, and
periodic boundary information (xst and xsc files), respectively. The
second group of files are restart files of coordinates, velocities, periodic
boundary information, and NAMD trajectory files (KL-EminEQ-I.dcd.),
respectively. Restart files are periodically updated by NAMD command,
restartfreq. More detailed information about config file and output file
can be found in the recent NAMD manual (http://
www.
ks.
uiuc.
edu/
Research/
namd/
).
3.1.3 Constrained MD Simulation
Once water and ion molecules are equilibrated at the target
temperature, it is necessary to apply another minimization and then
equilibrate the entire system while holding the KL complex with a weak
constraint. To generate a constraint file, load initial solvated system and
topology files (KL-Solvated.prmtop and KL-Solvated.inpcrd) to VMD as
described in Subheading 3.1.2. To assign constraints to the KL complex,
type the below commands to the VMD console.
vmd > set all [atomselect top all]
vmd > set ConstraintAtom [atomselect top "resname A C G U G5 C3"]
vmd > $all set beta 0
vmd > $ConstraintAtom set beta 0.5
vmd > $all writepdb KL-Constraint.pdb
Here, the beta column is set to zero, while atoms which belong to
the residue names, A, C, G, U, G5, and C3, are set to 0.5 kcal/(mol∙Å ). If a
biomolecular behavior is sensitive to equilibration, it may need to apply
strong initial constraint to the molecule and gradually reduce
constraints during multiple constrained MD simulations. The
constrained MD simulation can be performed by the below command.
namd2 +p number of cores EminEQ-II.conf > EminEQ-II.ene
The details of EminEQ-II.conf are listed in Table 1. Note that
coordinate, velocity, and periodic boundary files from the previous](https://image.slidesharecdn.com/111375-250318012721-6c51170a/75/RNA-Nanostructures-Design-Characterization-and-Applications-Methods-in-Molecular-Biology-2709-Kirill-A-Afonin-Editor-33-2048.jpg)
![simulation are specified as input files (line 2–6) in the EminEQ-II.conf.
In addition, the fixed KL complex is turned off (line 37), while the
constrained KL complex is activated (line 45). For the constrained MD
simulation, NVP is changed to NPT by turning off the initial boundary
box in line 54 and activating Langevin pressure control in line 71–81.
3.1.4 Final Equilibration and Product MD
Simulations
Once constrained MD simulation is completed, release constraints by
turning it off (line 45) in the EQ-III.conf file (see Table 1) to run the final
equilibration MD. This time, all components in the system, KL complex,
ions, and water, will be equilibrated at the target temperature. The
NAMD command for the final equilibration is below.
namd2 +p number of cores EQ-III.conf > EQ-III.ene
The details of the EQ-III.conf file are listed in Table 1. After the final
equilibration MD simulation is completed, the product MD simulation
can be performed using the below NAMD command.
namd2 +p number of cores MD.conf > MD.ene
The details of the MD.conf file are listed in Table 1. The MD.conf will
run to produce a 6 ns-long MD trajectory (3,000,000 step ×
2 fs = 6,000,000 fs = 6 ns). Longer MD trajectory can be produced by
running consecutive MD simulations using coordinate, velocity, and
periodic boundary files of the previous MD run. The results of MD
simulations are discussed in Note 2.
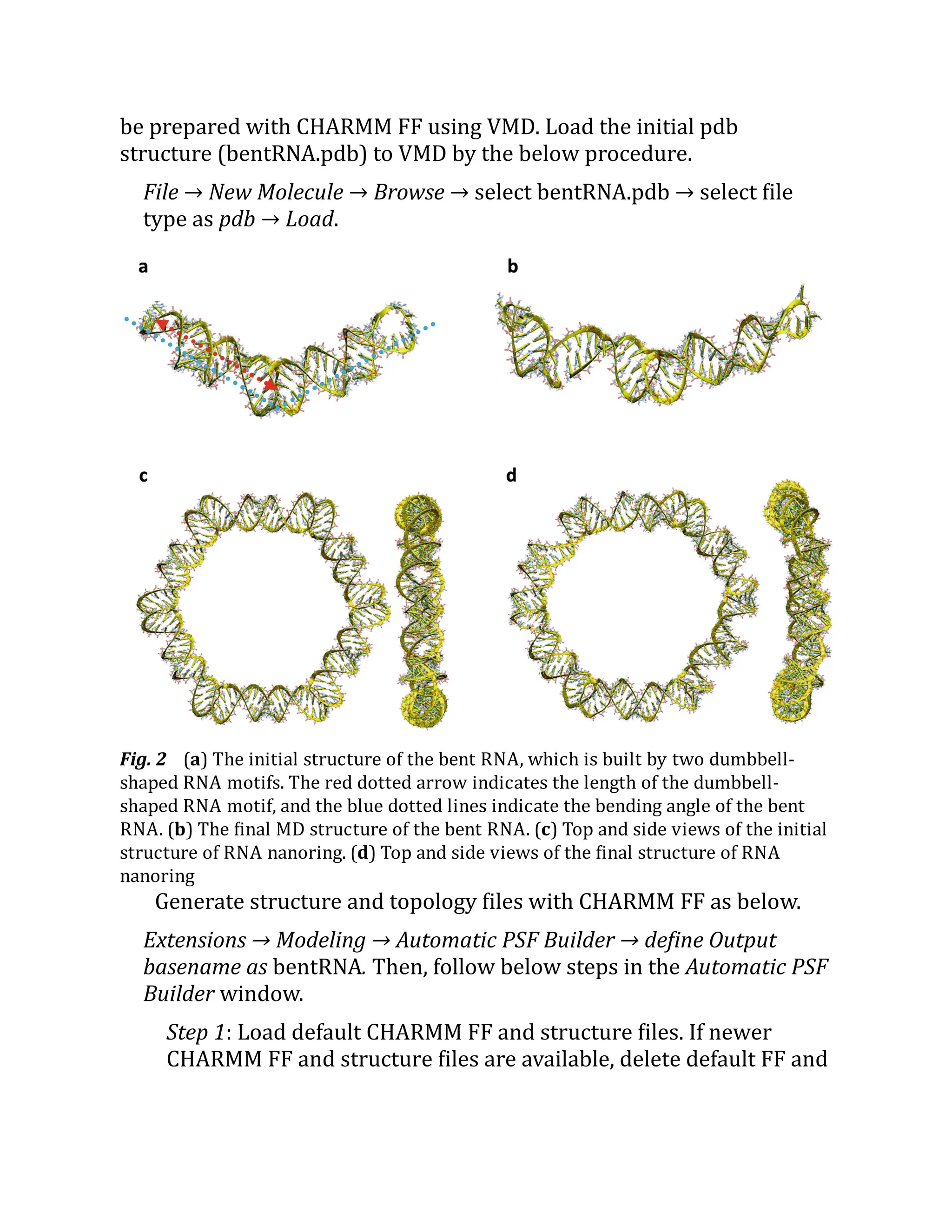
3.2 MD Simulation of a Bent RNA Motif and RNA
Nanoring
In this section, we introduce MD simulations of a bent RNA and an RNA
nanoring (Fig. 2). The bent RNA is composed by kissing loop
interactions between two dumbbell-shaped RNA motifs [11]. The RNA
nanostructure has a ring shape, which is constructed by six dumbbell-
shaped RNA motifs. Each dumbbell-shaped RNA has slightly different
sequences in the stem. The initial solvated structure and topology will](https://image.slidesharecdn.com/111375-250318012721-6c51170a/75/RNA-Nanostructures-Design-Characterization-and-Applications-Methods-in-Molecular-Biology-2709-Kirill-A-Afonin-Editor-34-2048.jpg)