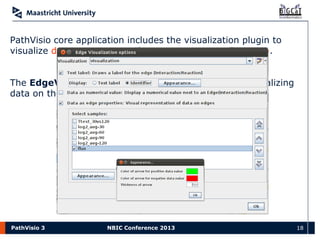
PathVisio 3 has been refactored using the OSGi framework, allowing it to be more modular and extensible through plugins. The new plugin system includes a plugin repository and manager that allows users to easily install plugins to add additional functionality. PathVisio's core functionality includes creating and editing pathways, visualizing experimental data on pathways, performing pathway analysis, and exporting results. It is open source and compatible with databases like Wikipathways, making it a flexible tool for pathway analysis.