This document serves as a comprehensive guide to parsing with Perl 6 grammars, introducing various coding constructs such as classes, roles, and grammars. It provides examples of how to define and use regex-based grammars for parsing different data formats, including FASTQ records and SQL translations. Additionally, it discusses specific grammar rules for handling SQL statements and translations to PostgreSQL syntax.









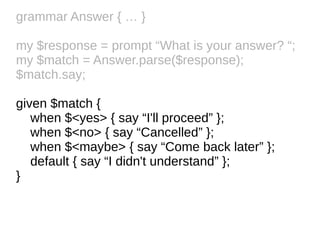
![Grammars are collections of named
regexes
grammar Answer {
regex TOP { <yes> | <no> | <maybe> }
regex yes { :i 'y' | 'ye' | 'yes' }
regex no { :i 'n' | 'no' }
regex maybe { :i
<{ 'maybe'.comb.produce(&[~]) }>
}
}](https://image.slidesharecdn.com/perl6-grammar-170414185144/85/Parsing-with-Perl6-Grammars-10-320.jpg)




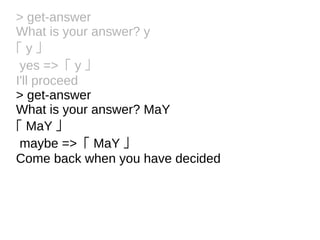
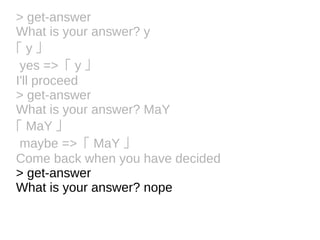
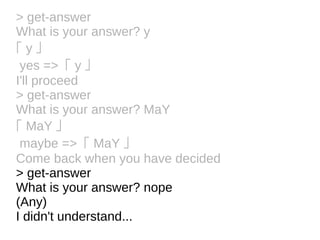



![Tokens and Rules
grammar Fastq {
…
token seq-id { .*? $$ }
token seq { <base>+ }
token base { <[A..Z] + [ - ] + [ * ]> }
token qual { <qual-letter>+ }
token qual-letter {
<[0..9 A..Z a..z ! " # $ % & ' ( ) * +
, - . / : ; < = > ?@ [ ] ^ _ ` { | } ~]>
}](https://image.slidesharecdn.com/perl6-grammar-170414185144/85/Parsing-with-Perl6-Grammars-22-320.jpg)


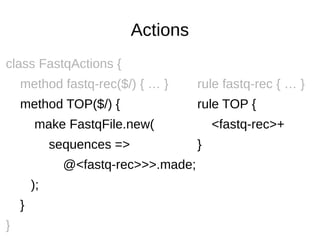





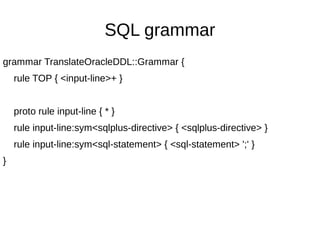
![sqlplus comments
grammar TranslateOracleDDL::Grammar {
rule input-line:sym<sqlplus-directive> { <sqlplus-directive> }
token string-to-end-of-line { V+ }
rule sqlplus-directive:sym<REM> {
['REM'<?before v>]
| ['REM'<string-to-end-of-line>
}
rule sqlplus-directive:sym<PROMPT> {
['PROMPT'<?before v>]
| ['PROMPT'<string-to-end-of-line>]
}
}](https://image.slidesharecdn.com/perl6-grammar-170414185144/85/Parsing-with-Perl6-Grammars-32-320.jpg)
![Translation Actions
grammar TranslateOracleDDL::Grammar {
rule sqlplus-directive:sym<REM> {
['REM'<?before v>]
| ['REM'<string-to-end-of-line>
}
}
class TranslateOracleDDL::ToPostgres {
method sql-statement:sym<REM>($/) {
make '--' ~ ($<string-to-end-of-line> || '' )
}
}](https://image.slidesharecdn.com/perl6-grammar-170414185144/85/Parsing-with-Perl6-Grammars-33-320.jpg)



![SQL statements
grammar TranslateOracleDDL::Grammar {
rule input-line:sym<sql-statement> { <sql-statement> ';' }
proto rule sql-statement { * }
rule sql-statement:sym<CREATE-SEQUENCE> { … }
rule sql-statement:sym<COMMENT-ON> {
'COMMENT' 'ON' ['TABLE' | 'COLUMN'] <entity-name> IS <value>
}
}](https://image.slidesharecdn.com/perl6-grammar-170414185144/85/Parsing-with-Perl6-Grammars-37-320.jpg)
![SQL statements
grammar TranslateOracleDDL::Grammar {
rule input-line:sym<sql-statement> { <sql-statement> ';' }
proto rule sql-statement { * }
rule sql-statement:sym<CREATE-SEQUENCE> { … }
rule sql-statement:sym<COMMENT-ON> { … }
rule sql-statement:sym<CREATE-TABLE> {
'CREATE' 'TABLE' <entity-name>
'(' <create-table-column-def>+? % ','
[ ',' <table-constraint-def> ]*
')'
<create-table-extra-oracle-stuff>*
}](https://image.slidesharecdn.com/perl6-grammar-170414185144/85/Parsing-with-Perl6-Grammars-38-320.jpg)
![SQL statements
grammar TranslateOracleDDL::Grammar {
rule input-line:sym<sql-statement> { <sql-statement> ';' }
proto rule sql-statement { * }
rule sql-statement:sym<CREATE-SEQUENCE> { … }
rule sql-statement:sym<COMMENT-ON> { … }
rule sql-statement:sym<CREATE-TABLE> { … }
rule sql-statement:sym<CREATE-INDEX> {
'CREATE' [ $<unique>=('UNIQUE') ]?
'INDEX' <index-name=entity-name>
'ON' <table-name=entity-name>
'(' ~ ')' <columns=expr>+ % ','
<index-option>*](https://image.slidesharecdn.com/perl6-grammar-170414185144/85/Parsing-with-Perl6-Grammars-39-320.jpg)
![SQL statements
grammar TranslateOracleDDL::Grammar {
rule input-line:sym<sql-statement> { <sql-statement> ';' }
proto rule sql-statement { * }
rule sql-statement:sym<CREATE-SEQUENCE> { … }
rule sql-statement:sym<COMMENT-ON> { … }
rule sql-statement:sym<CREATE-TABLE> { … }
rule sql-statement:sym<CREATE-INDEX> { … }
rule sql-statement:sym<CREATE-VIEW> {
'CREATE' [ 'OR' 'REPLACE' ]? 'VIEW' <view-name=entity-name>
'(' ~ ')' <columns=expr> + % ','
'AS' <select-statement>
}](https://image.slidesharecdn.com/perl6-grammar-170414185144/85/Parsing-with-Perl6-Grammars-40-320.jpg)
![Table, Column, Sequence, etc.
names
grammar TranslateOracleDDL::Grammar {
rule entity-name { <identifier>** 1..3 % '.' }
proto token identifier { * }
token identifier:sym<bareword> { <[$w]>+ }
token identifier:sym<qq> {
'”'
[ <-[“]>+:
| '””'
]*
'”'
}](https://image.slidesharecdn.com/perl6-grammar-170414185144/85/Parsing-with-Perl6-Grammars-41-320.jpg)










![SQL expressions
grammar TranslateOracleDDL::Grammar {
token and-or { :i 'and' | 'or' }
proto rule expr { * }
rule expr:sym<nested> { [ '(' <expr> ')' ]+ % <and-or> }
rule expr:sym<composite> { <expr> <and-or> <expr> }
rule expr:sym<atom> { <entity-name> | <value> }
rule expr:sym<operator> { <left=identifier-or-value>
<expr-op>
<right=expr> }
rule expr:sym<IN> { 'IN' '(' ~ ')' <value> + % ',' }
rule expr:sym<substr-f> { 'substr(' <expr>**2..3 % ',' ')' }](https://image.slidesharecdn.com/perl6-grammar-170414185144/85/Parsing-with-Perl6-Grammars-52-320.jpg)
![Left-recursive grammar
grammar TranslateOracleDDL::Grammar {
token and-or { :i 'and' | 'or' }
proto rule expr { * }
rule expr:sym<nested> { [ '(' <expr> ')' ]+ % <and-or> }
rule expr:sym<composite> { <expr> <and-or> <expr> }
rule expr:sym<atom> { <entity-name> | <value> }
rule expr:sym<operator> { <left=ident-or-value>
<expr-op>
<right=expr> }
rule expr:sym<IN> { 'IN' '(' ~ ')' <value> + % ',' }
rule expr:sym<substr-f> { 'substr(' <expr>**2..3 % ',' ')' }](https://image.slidesharecdn.com/perl6-grammar-170414185144/85/Parsing-with-Perl6-Grammars-53-320.jpg)
![Remove left recursion
grammar TranslateOracleDDL::Grammar {
token and-or { :i 'and' | 'or' }
proto rule expr { * }
rule expr:sym<nested> { [ '(' <expr> ')' ]+ % <and-or> }
rule expr:sym<composite> { <expr-part> <and-or> <expr> }
rule expr:sym<part> { <expr-part> }
rule expr:sym<atom> { <entity-name> | <value> }
rule expr-part:sym<operator> { <left=ident-or-value>
<expr-op>
<right=expr> }
rule expr-part:sym<IN> { 'IN' '(' ~ ')' <value> + % ',' }
rule expr-part:sym<substr-f> { 'substr(' <expr>**2..3 % ',' ')' }](https://image.slidesharecdn.com/perl6-grammar-170414185144/85/Parsing-with-Perl6-Grammars-54-320.jpg)
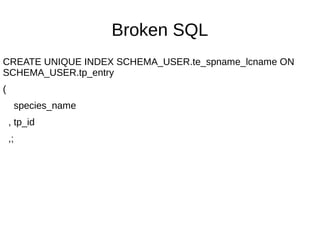
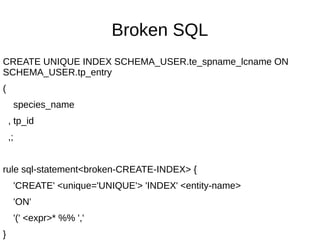

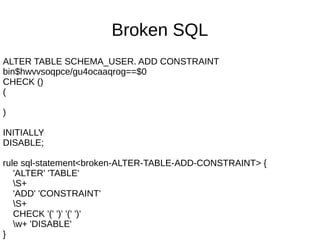
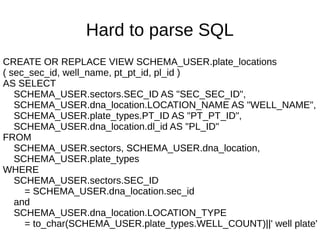

![Hard to parse SQL
rule sql-statement:sym<special-CREATE-VIEW> {
'CREATE' 'OR' 'REPLACE' 'VIEW'
'SCHEMA_USER.plate_locations'
'(' <-[;]>+
}
method sql-statement:sym<special-CREATE-VIEW>($/) {
Make ~$/;
}](https://image.slidesharecdn.com/perl6-grammar-170414185144/85/Parsing-with-Perl6-Grammars-61-320.jpg)
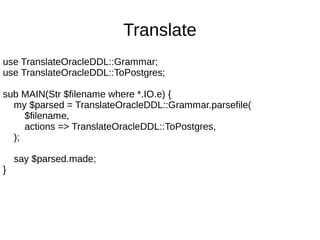

![Debugging
rule select-statement { :ignorecase
'SELECT'
[ $<distinct>=('DISTINCT') ]?
<columns=select-column>+ % ','
'FROM'
<select-from-clause> + % ','
<join-clause>*
<where-clause>?
<group-by-clause>?
}](https://image.slidesharecdn.com/perl6-grammar-170414185144/85/Parsing-with-Perl6-Grammars-64-320.jpg)
![Debugging
rule select-statement { :ignorecase
'SELECT' { say “saw SELECT” }
[ $<distinct>=('DISTINCT') { say “saw DISTINCT” }]?
<columns=select-column>+ % ',' { say @<columns>.elems,” columns” }
'FROM'
<select-from-clause> + % ',' { say @<select-from-
clause>.elems,”from”}
<join-clause>* { say @<join-clause>.elems, “ joins” }
<where-clause>?
<group-by-clause>?
}](https://image.slidesharecdn.com/perl6-grammar-170414185144/85/Parsing-with-Perl6-Grammars-65-320.jpg)
