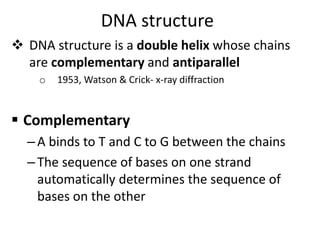
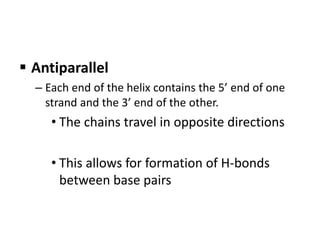
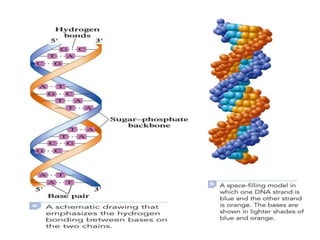
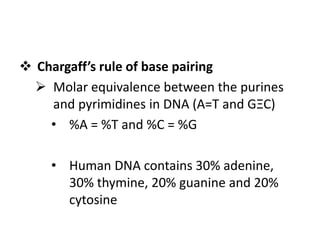
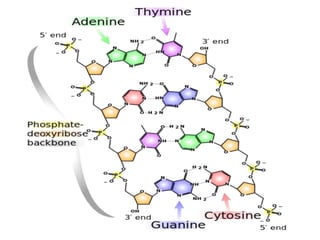
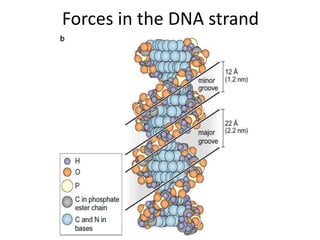
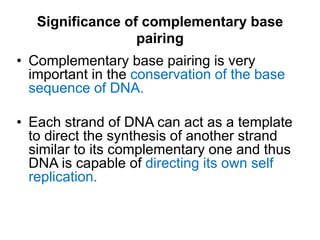
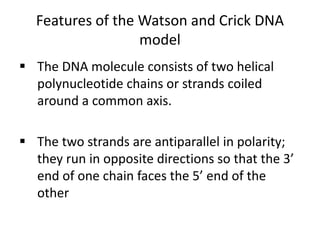
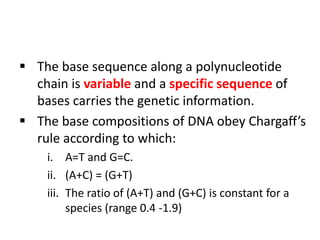
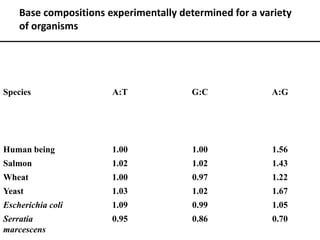
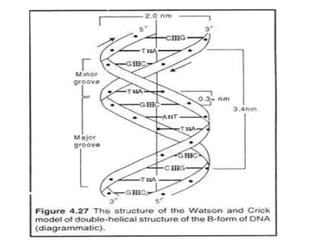
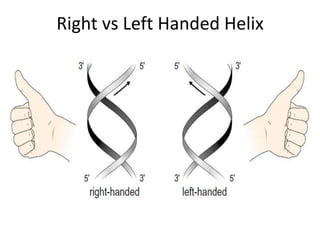
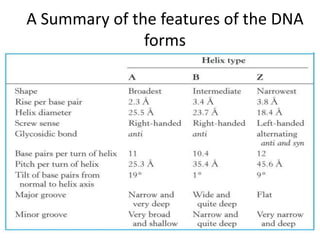
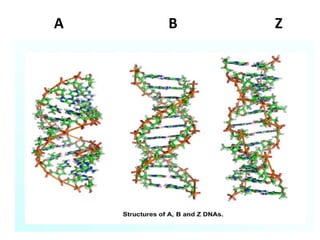
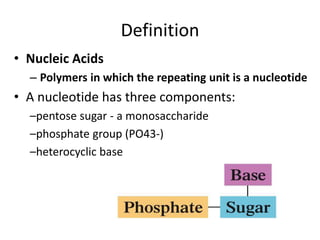
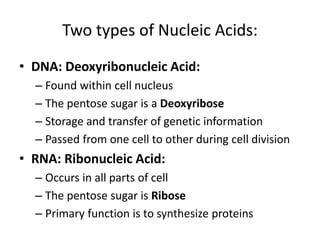
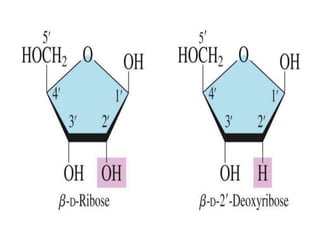
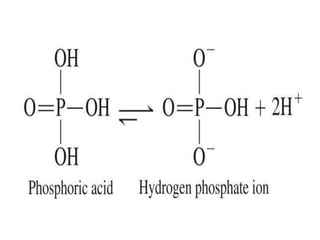
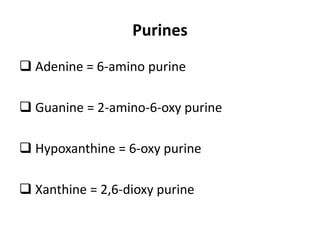
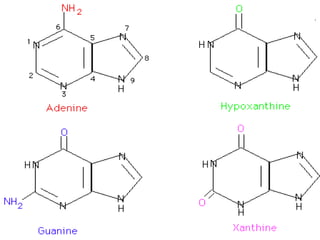
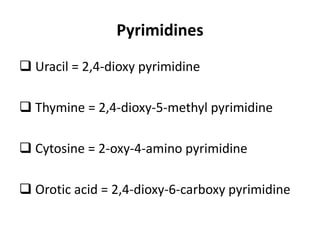
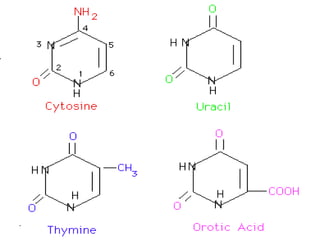
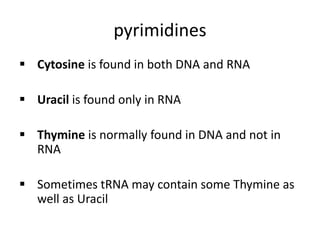
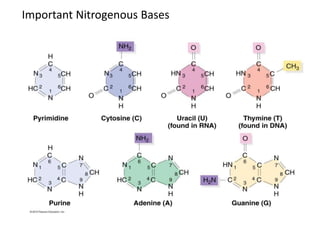
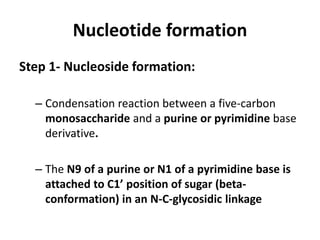
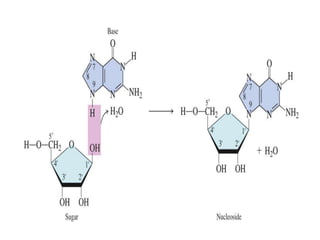
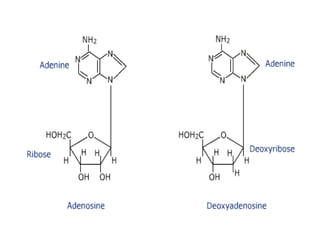
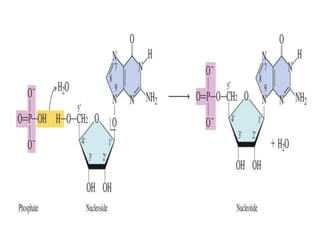
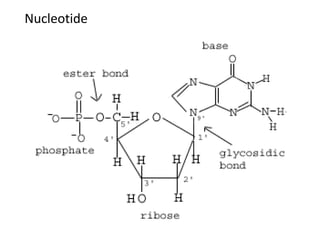
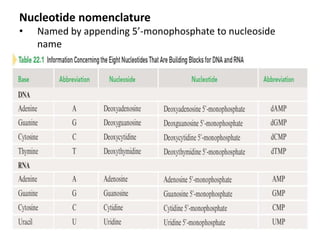
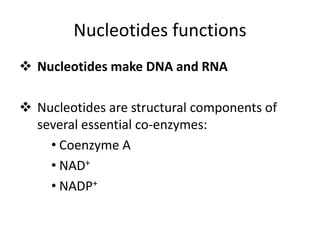
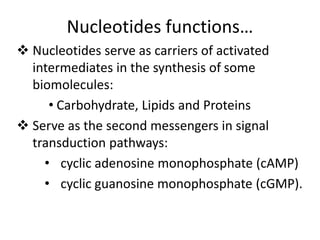
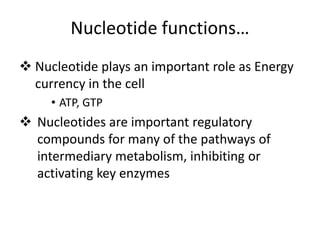
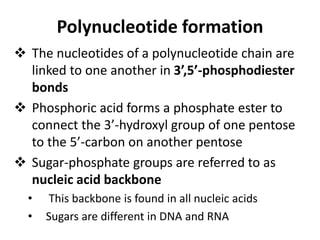
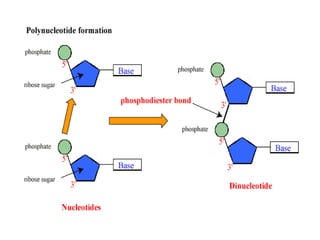
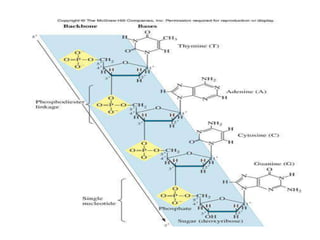
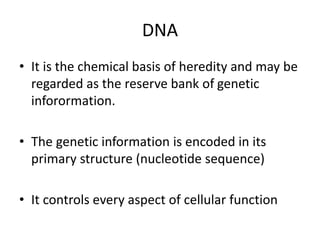
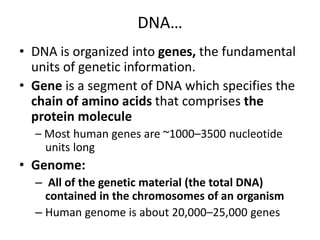
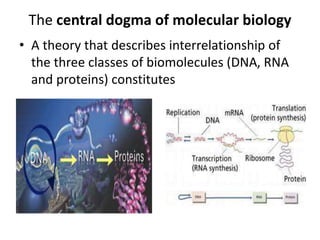
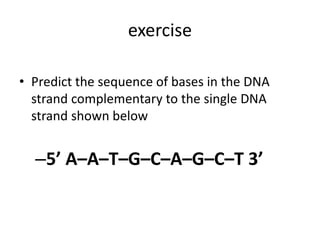Nucleic acids are polymers made of nucleotides that serve as the repository of genetic information. There are two main types of nucleic acids: DNA and RNA. DNA is found in cell nuclei and contains the genetic blueprint. RNA is found throughout the cell and assists in protein synthesis. A nucleotide contains a nitrogenous base (purine or pyrimidine), a 5-carbon sugar (deoxyribose in DNA and ribose in RNA), and one or more phosphate groups. Nucleotides bond together via phosphodiester linkages between the sugar and phosphate to form polynucleotide chains. DNA exists as a double helix with the bases on the inside bonded via hydrogen bonds in a complementary and antiparallel fashion







![DNA molecules from higher organisms can be
much larger
The human genome comprises of 3 billion
nucleotides [3 x 109], divided among 23 distinct
DNA molecules (chromosomes) of different
sizes.
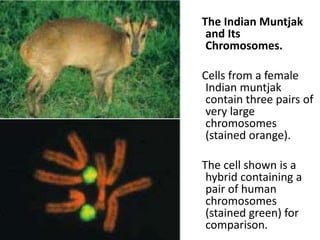
One of the largest known DNA molecules is
found in the Indian muntjak, an Asiatic deer; its
genome is nearly as large as the human genome
but is distributed on only 3 chromosomes](https://image.slidesharecdn.com/nucleicacidschasama-230103165404-2a2dfca3/85/NUCLEIC-ACIDS-chasama-pptx-34-320.jpg)