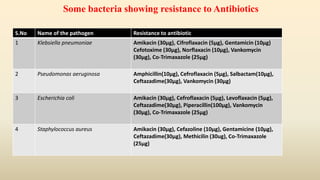
The document details a study focused on collecting and analyzing hospital sewage and municipal drain samples to identify multidrug resistant bacteria and the influence of urbanization on antibiotic resistance. It outlines the processes of sample collection, bacterial isolation, antibiotic sensitivity testing, and molecular identification of specific pathogens. Key findings included the presence of multidrug-resistant strains such as Klebsiella pneumoniae and Pseudomonas aeruginosa, which pose significant public health risks due to their resistance to multiple antibiotics.