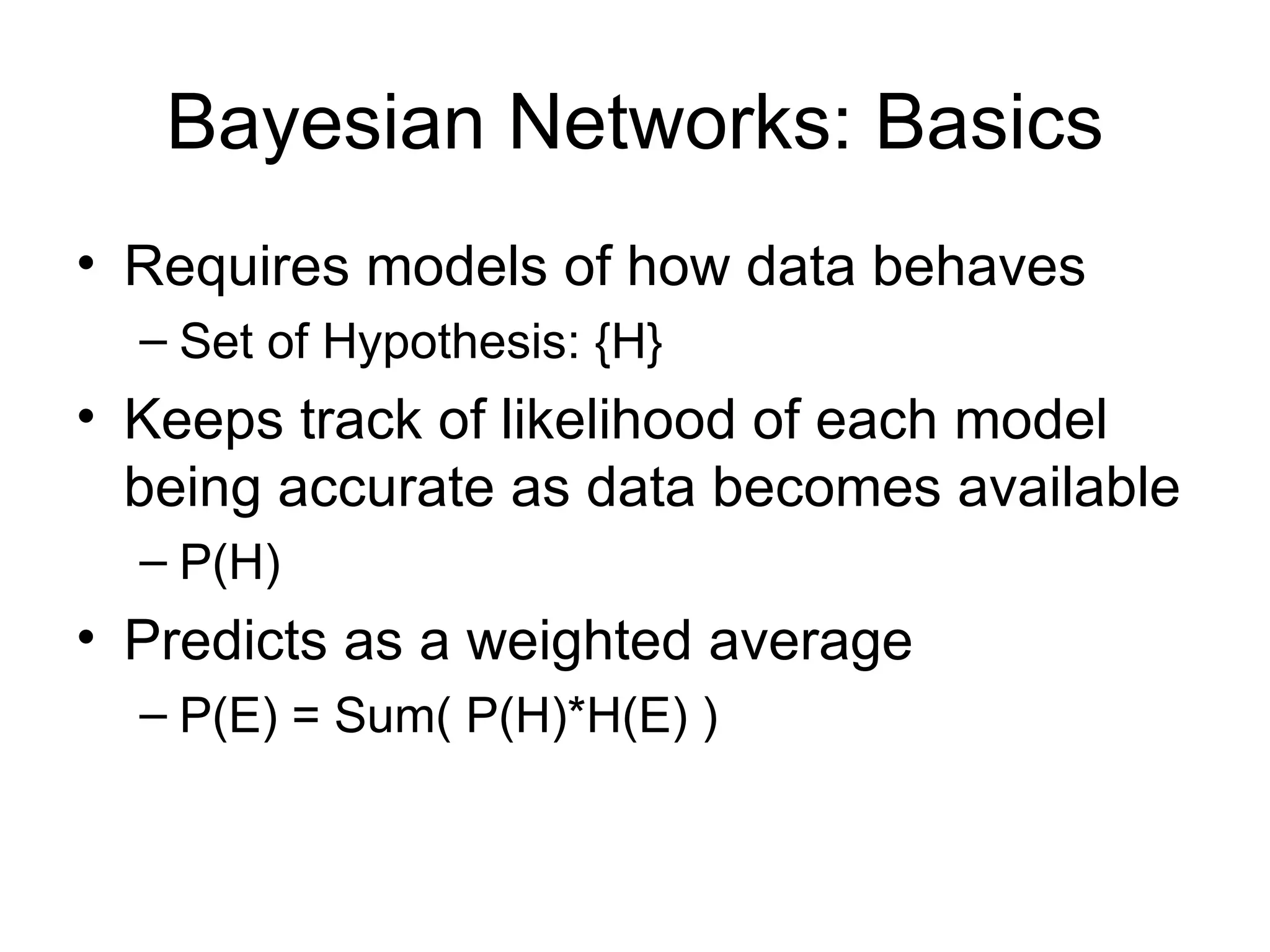
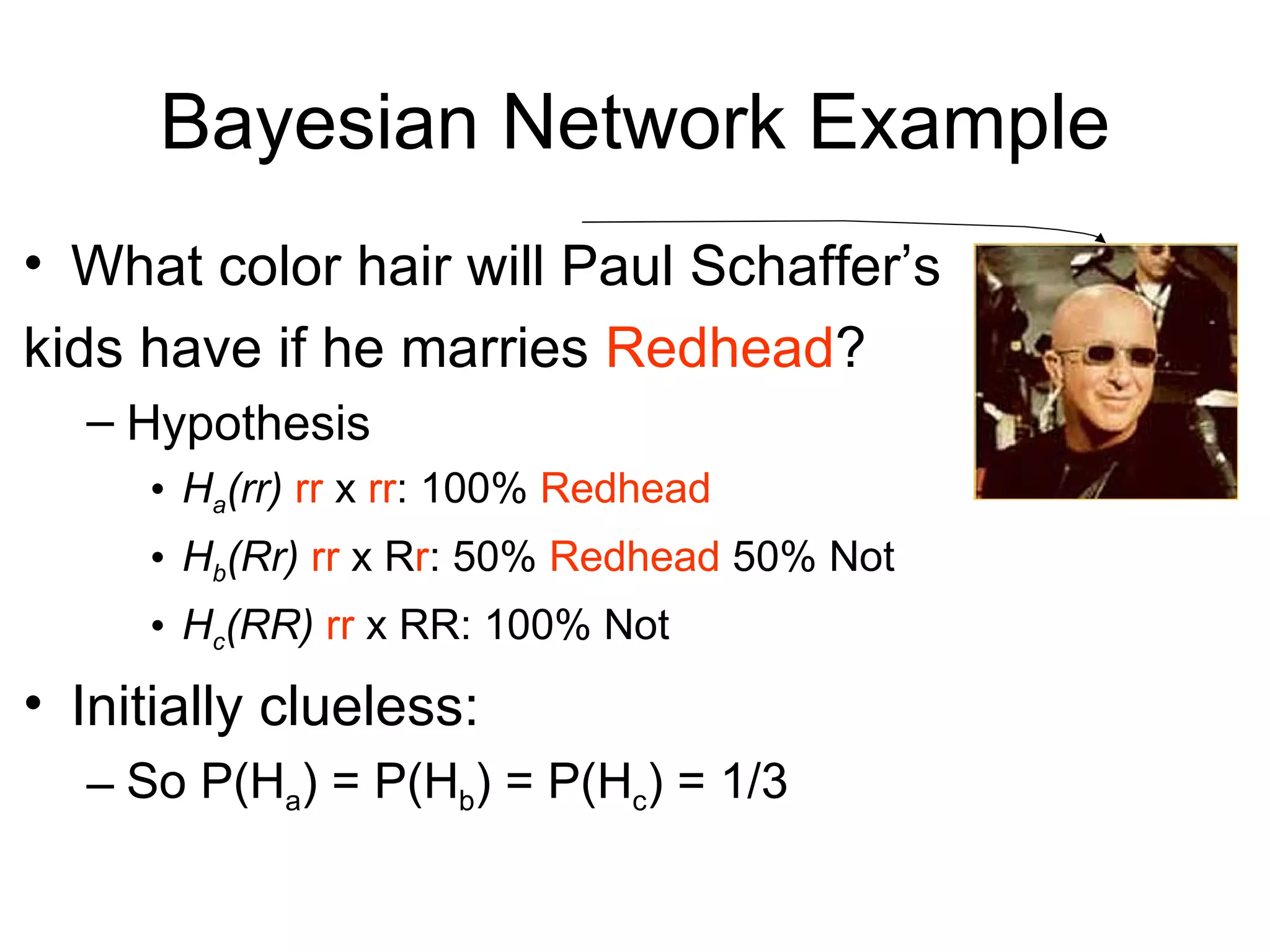
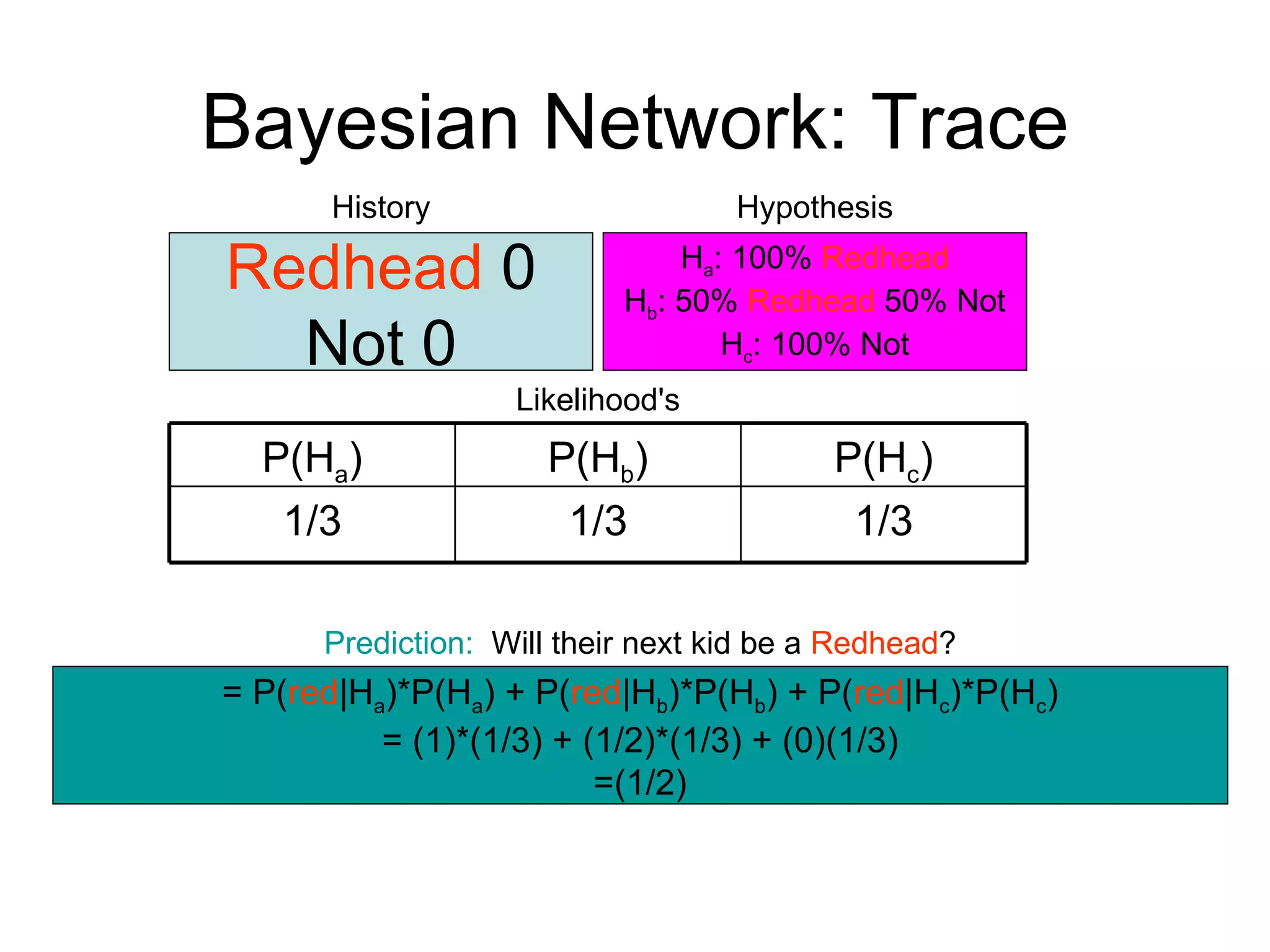
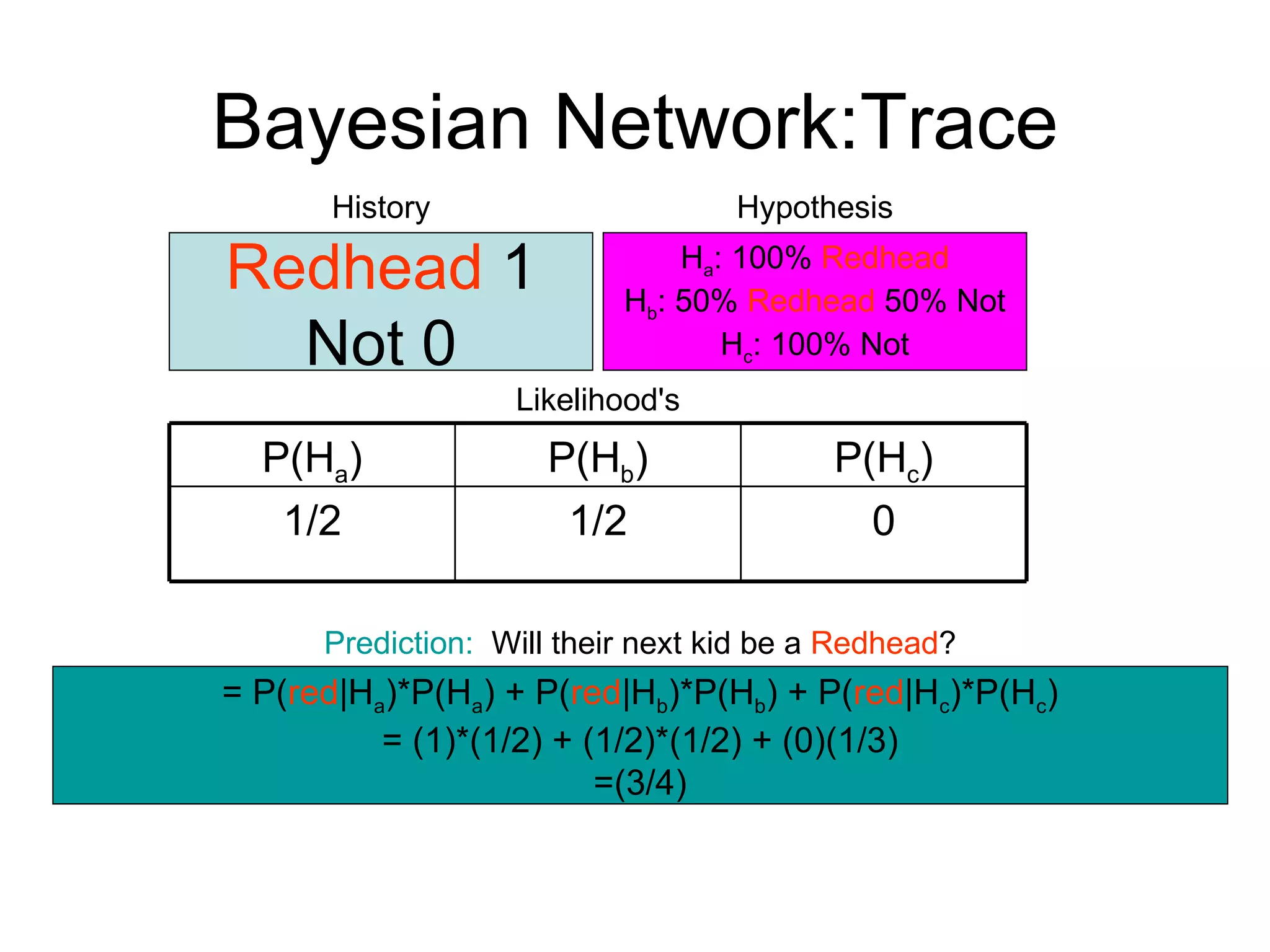
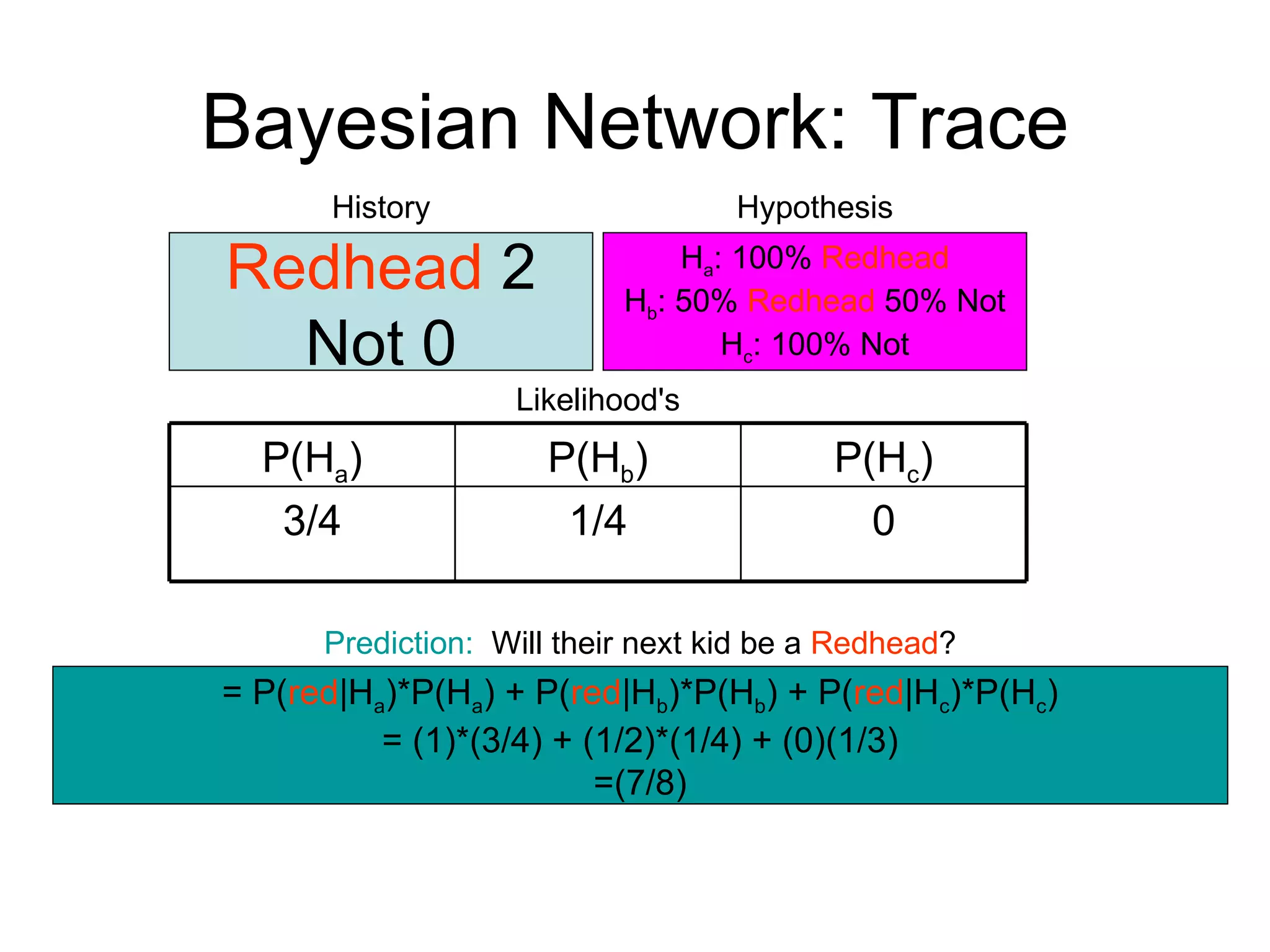
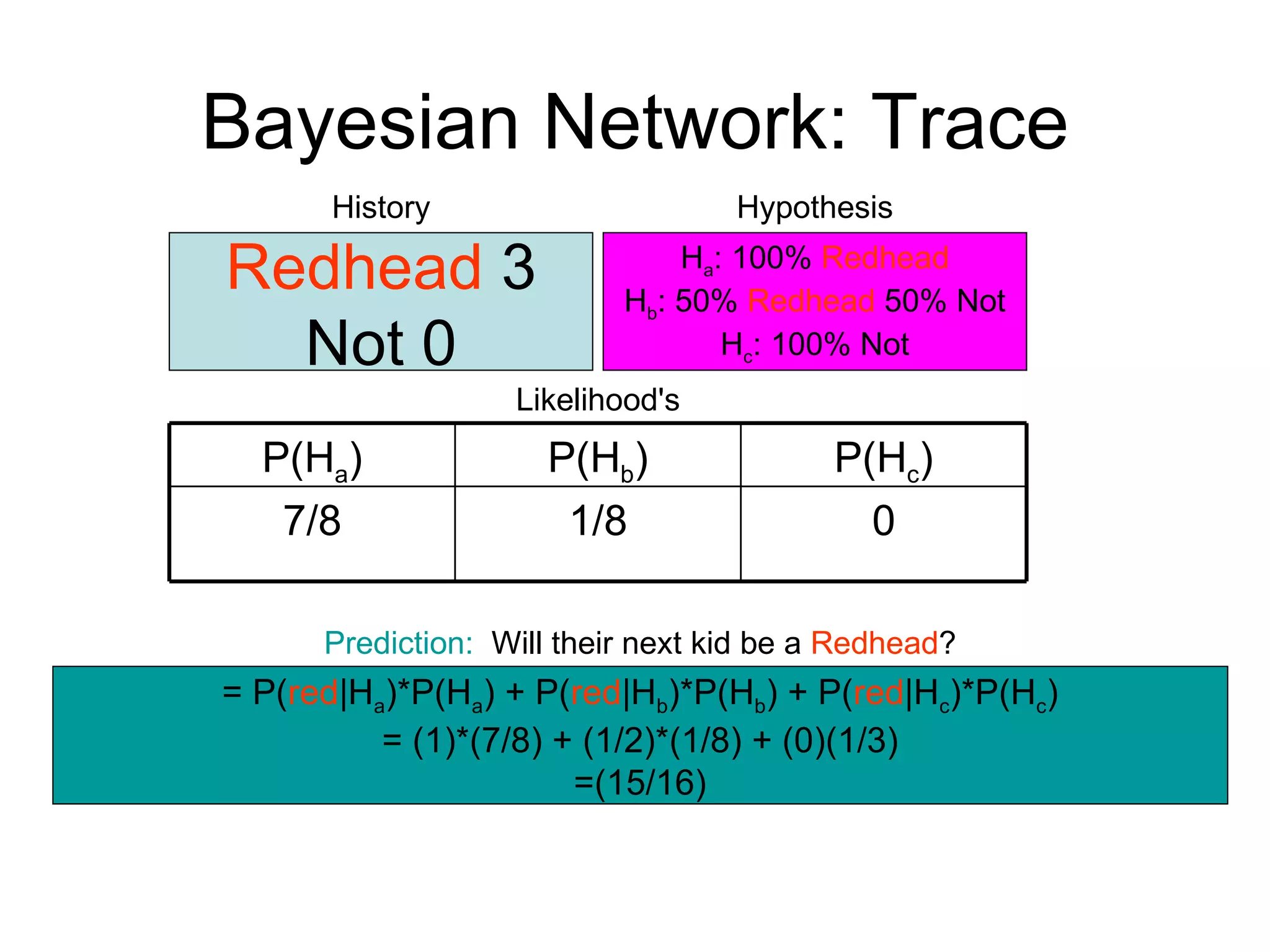
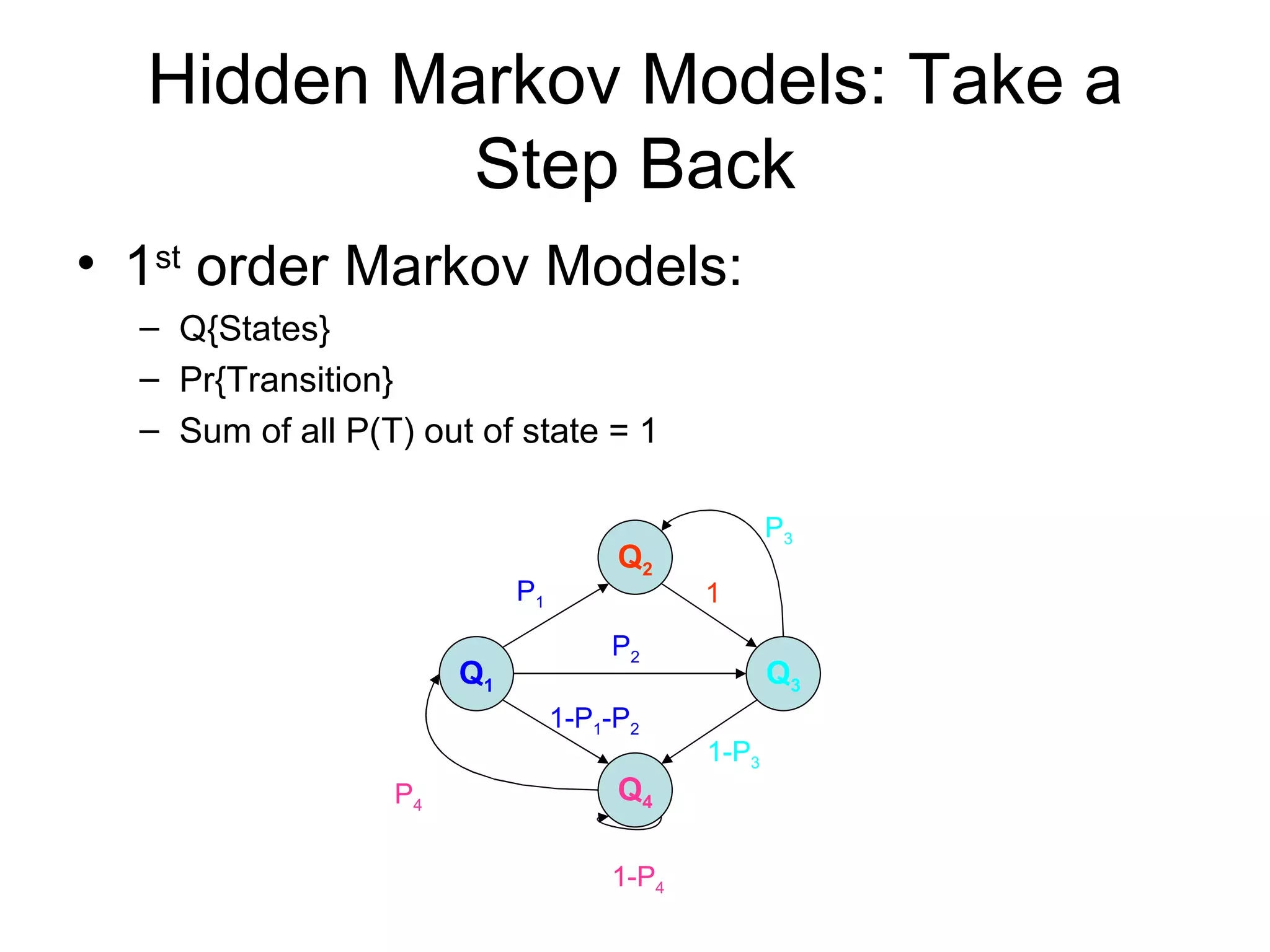
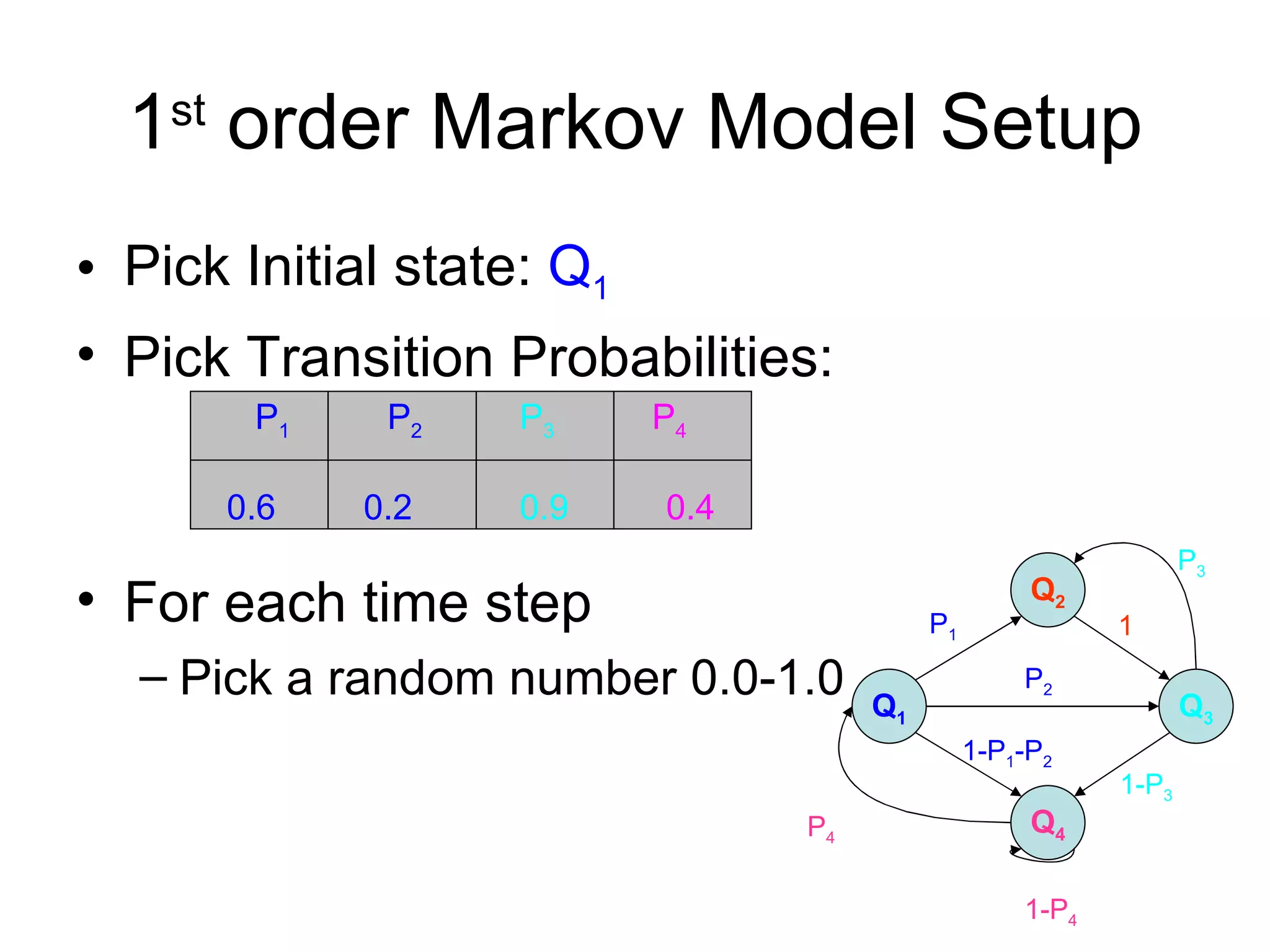
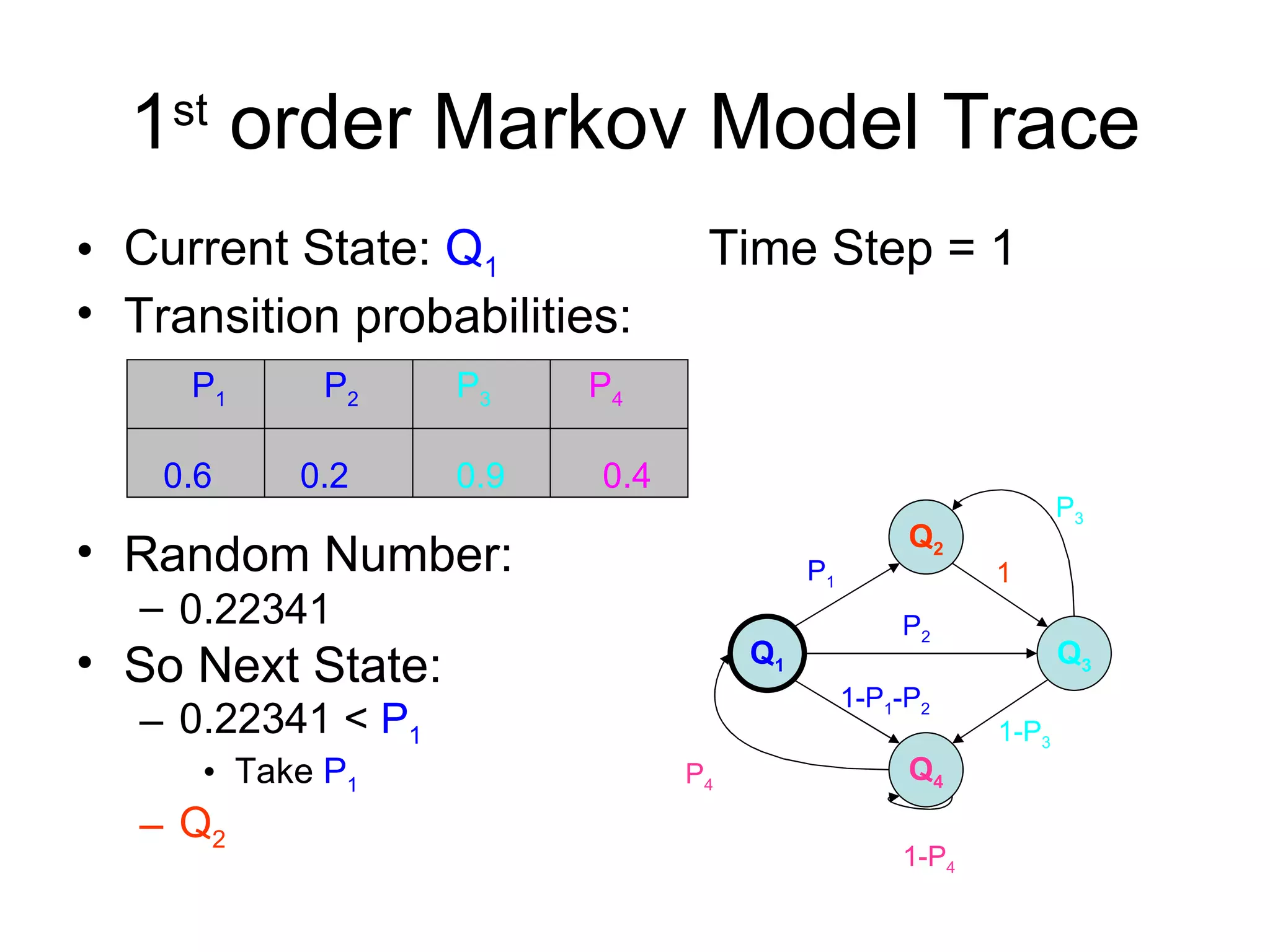
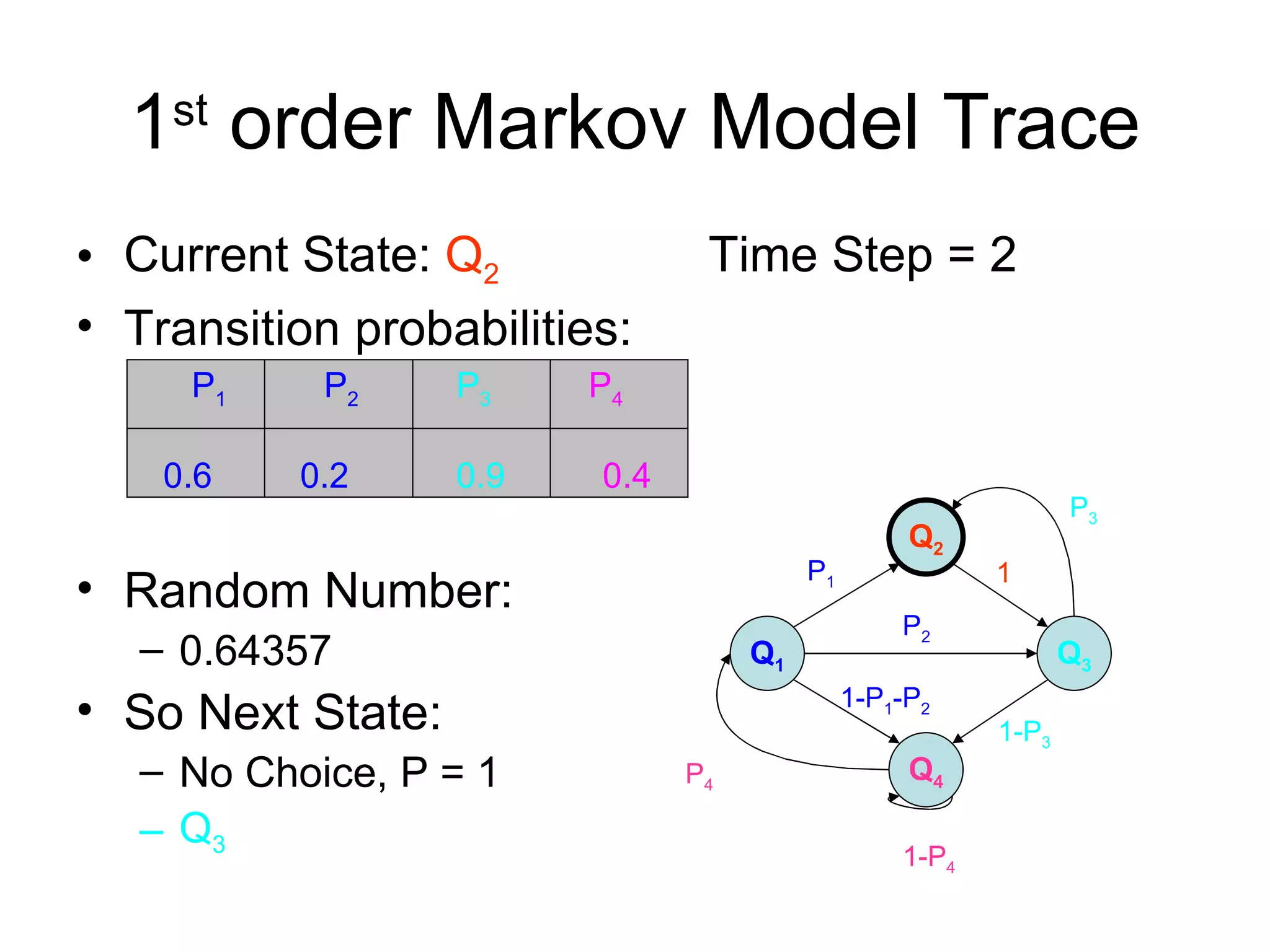
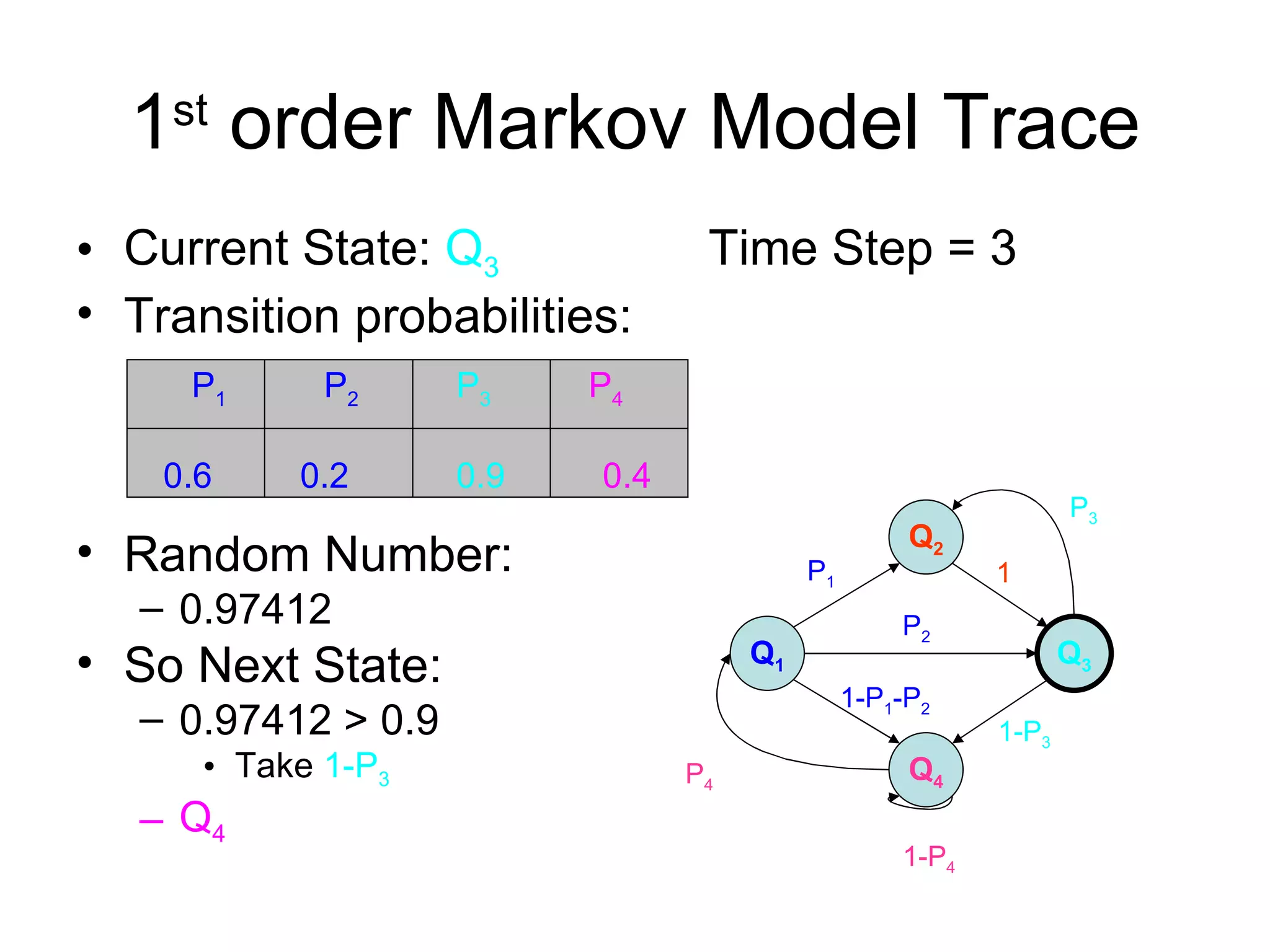
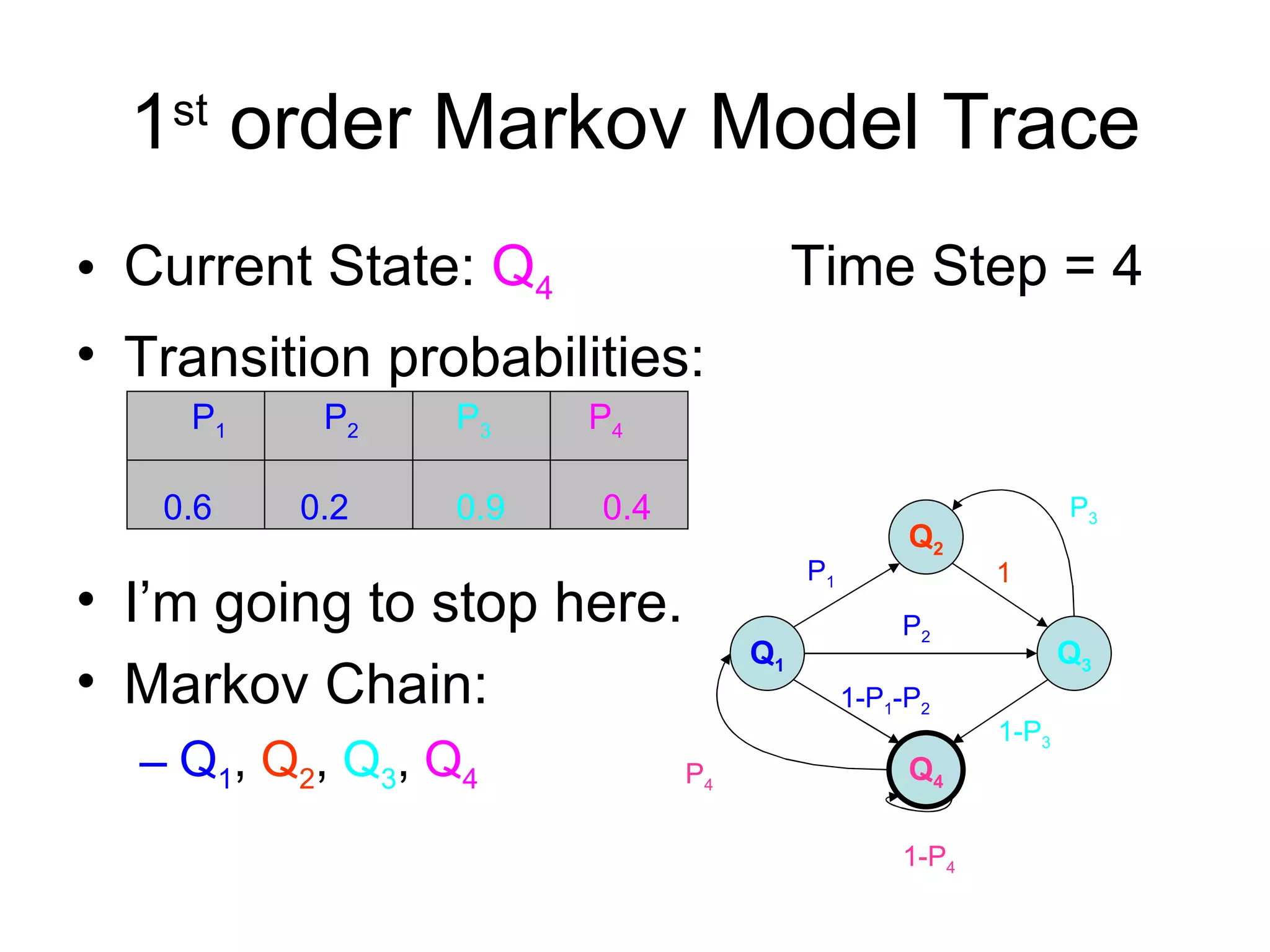
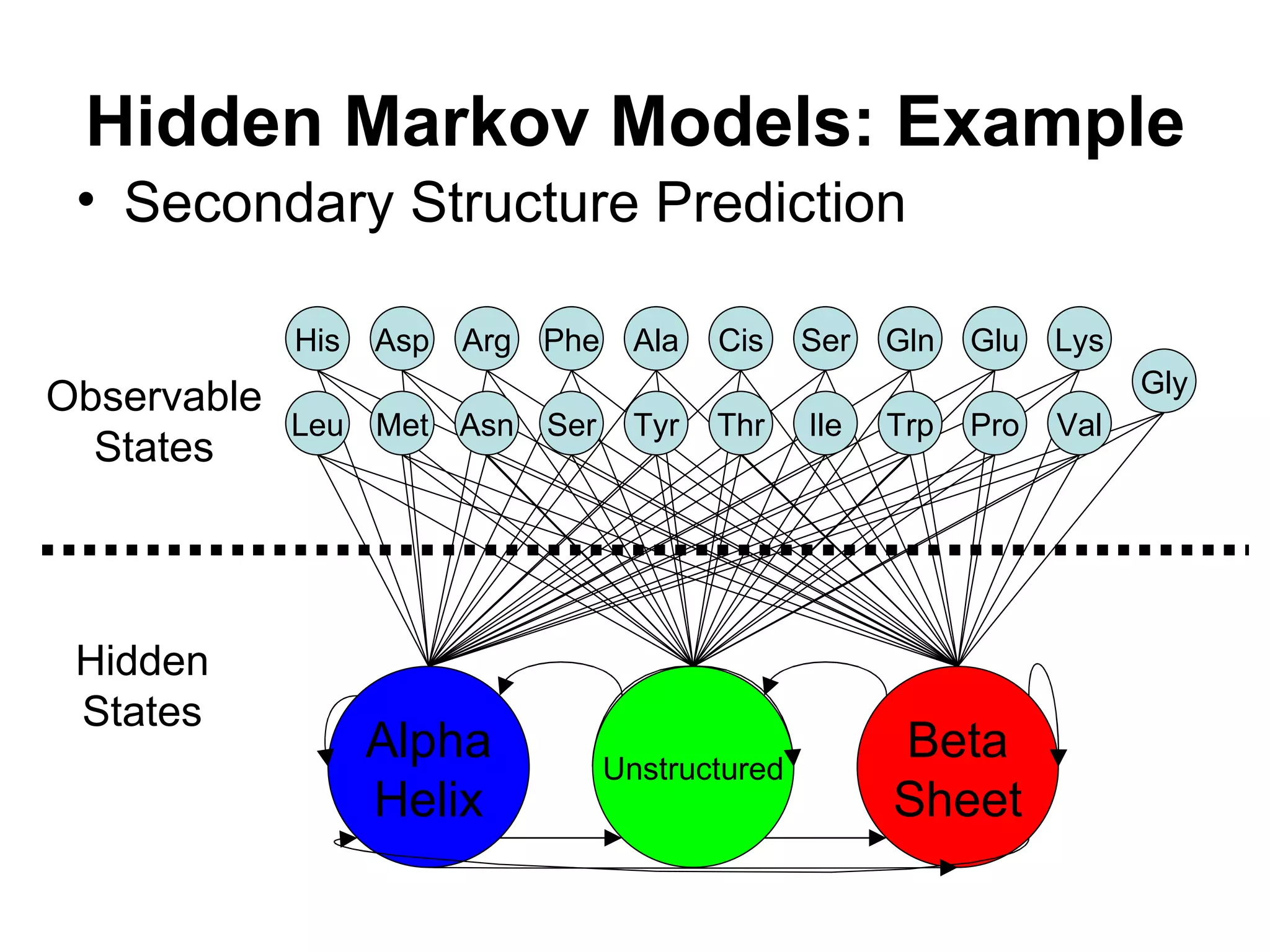
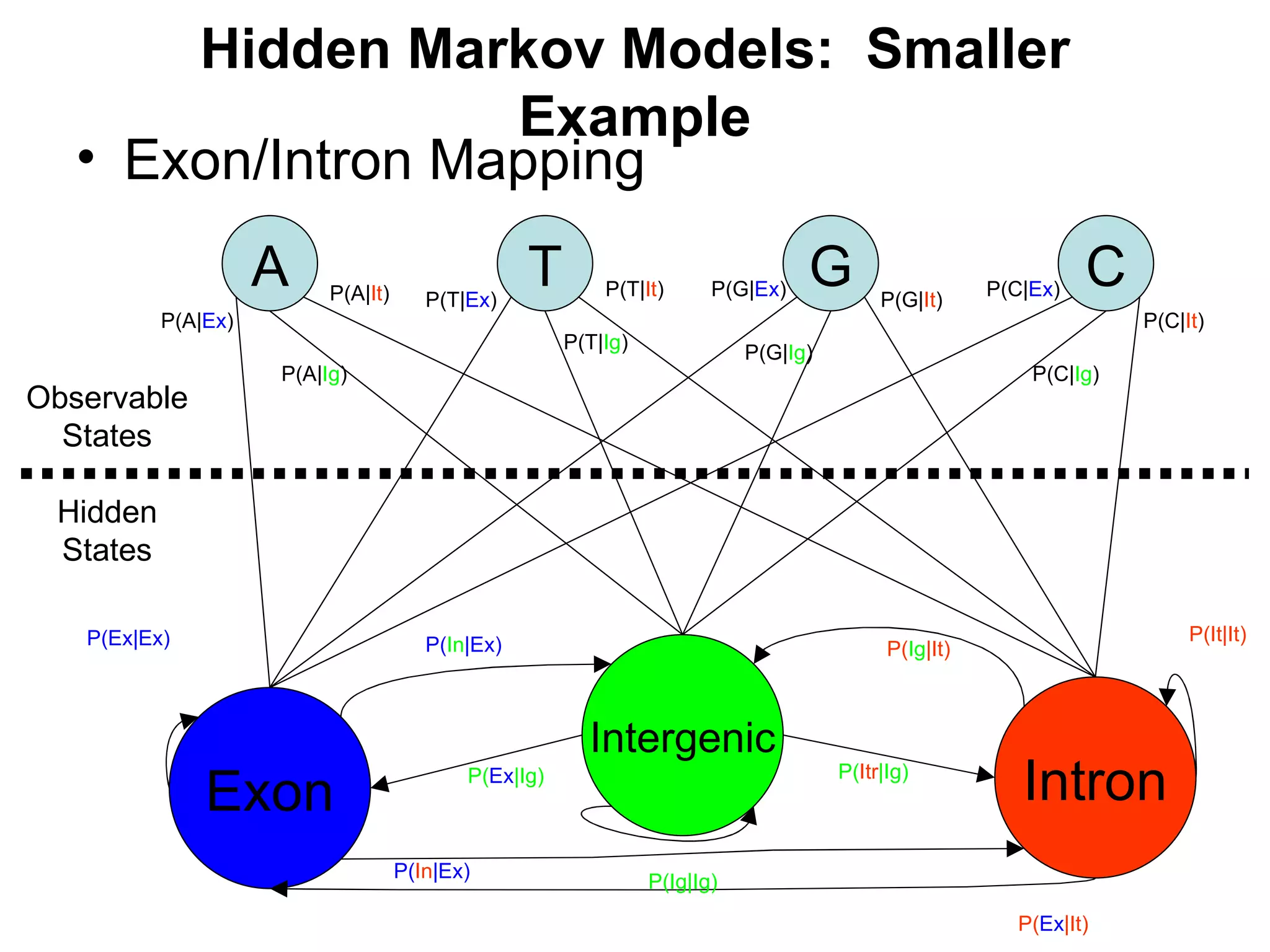
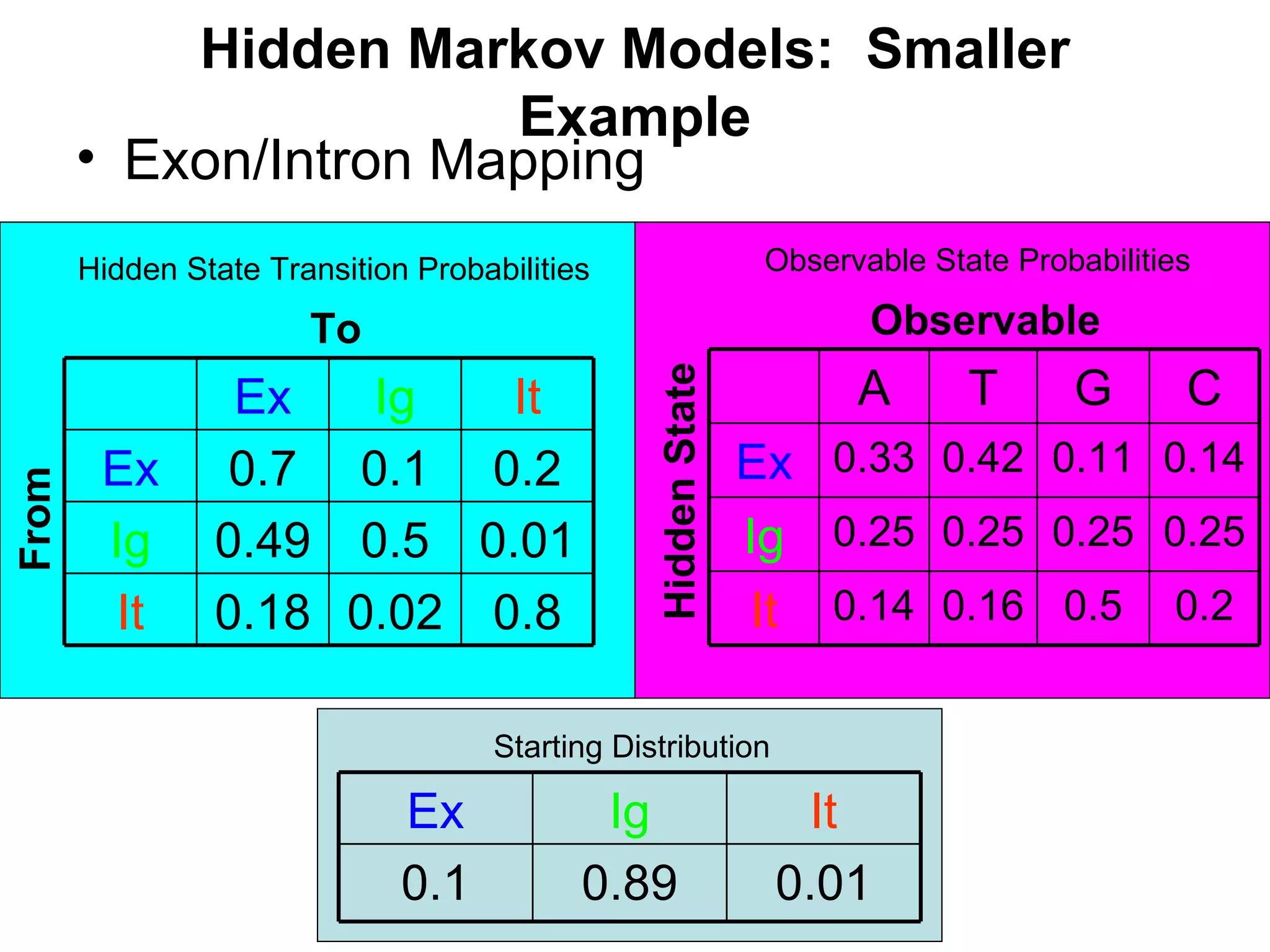
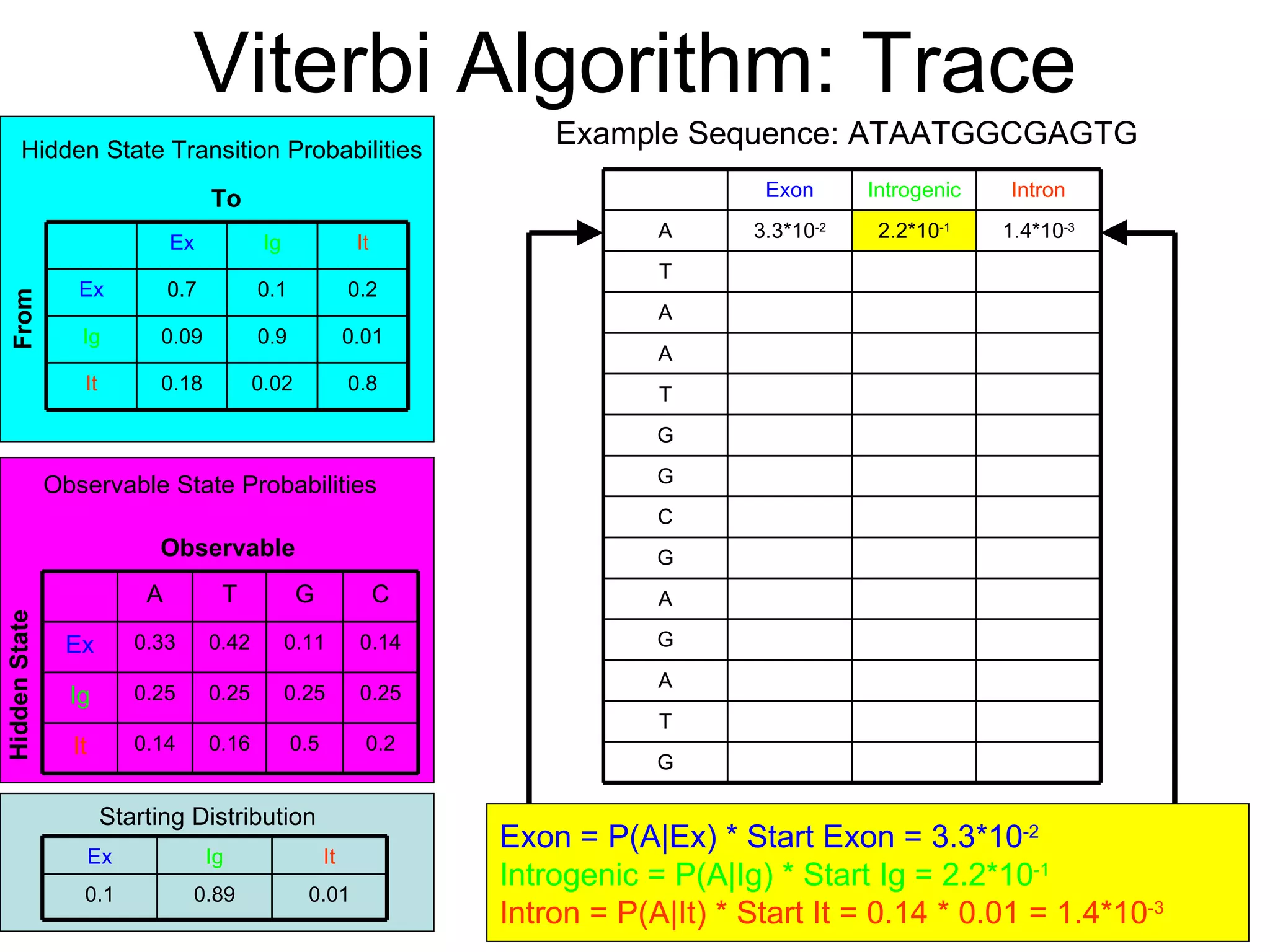
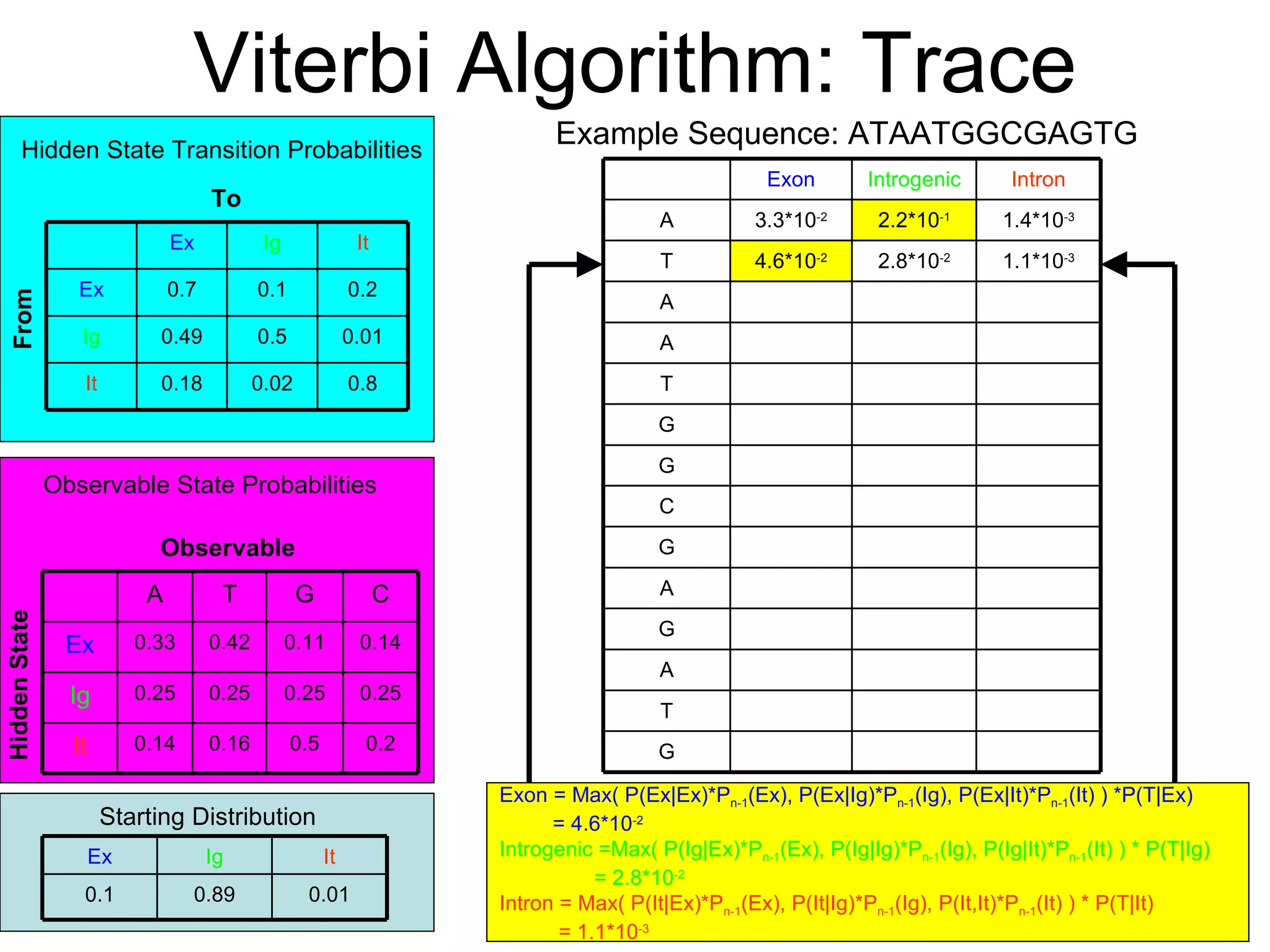
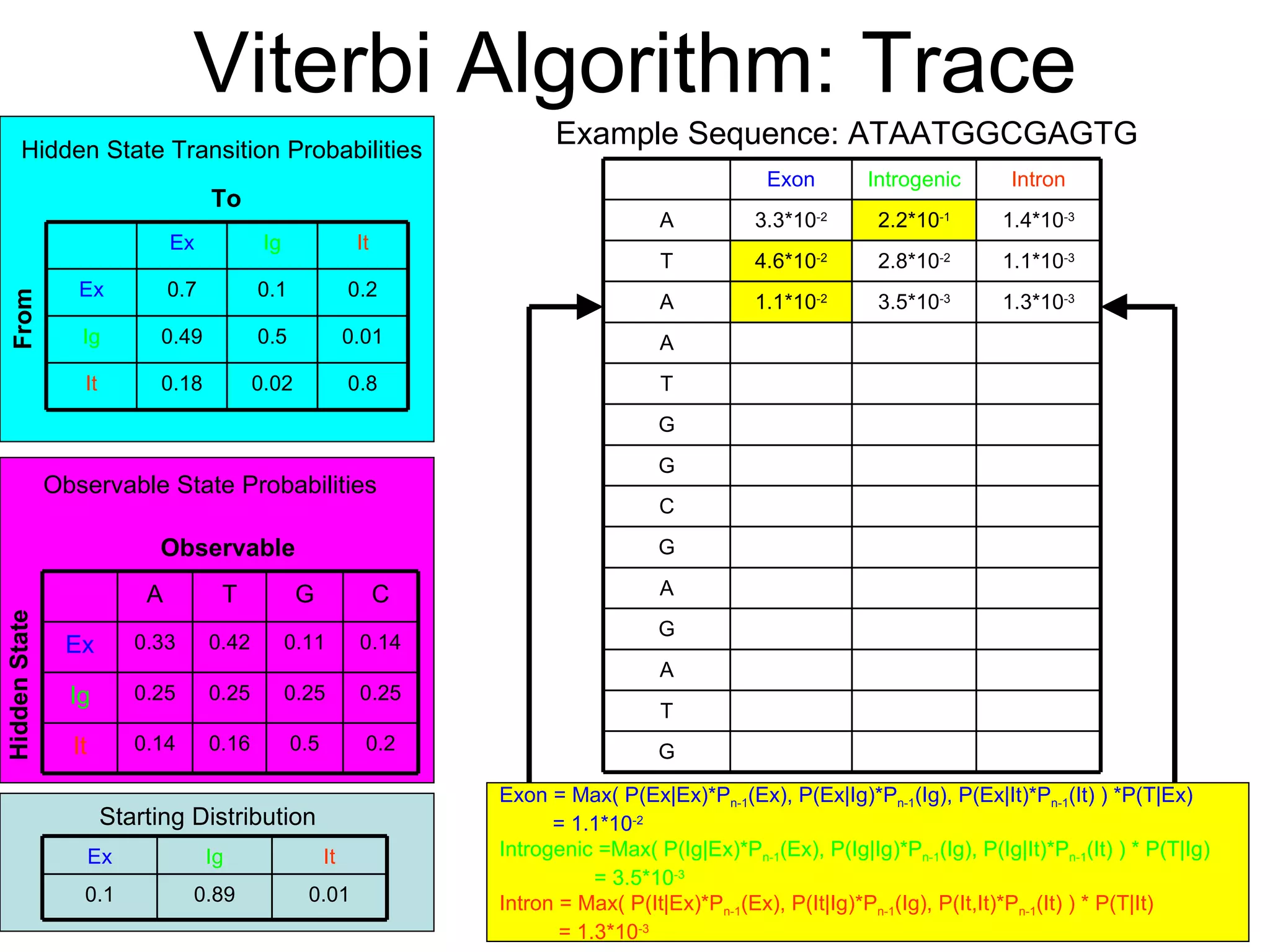
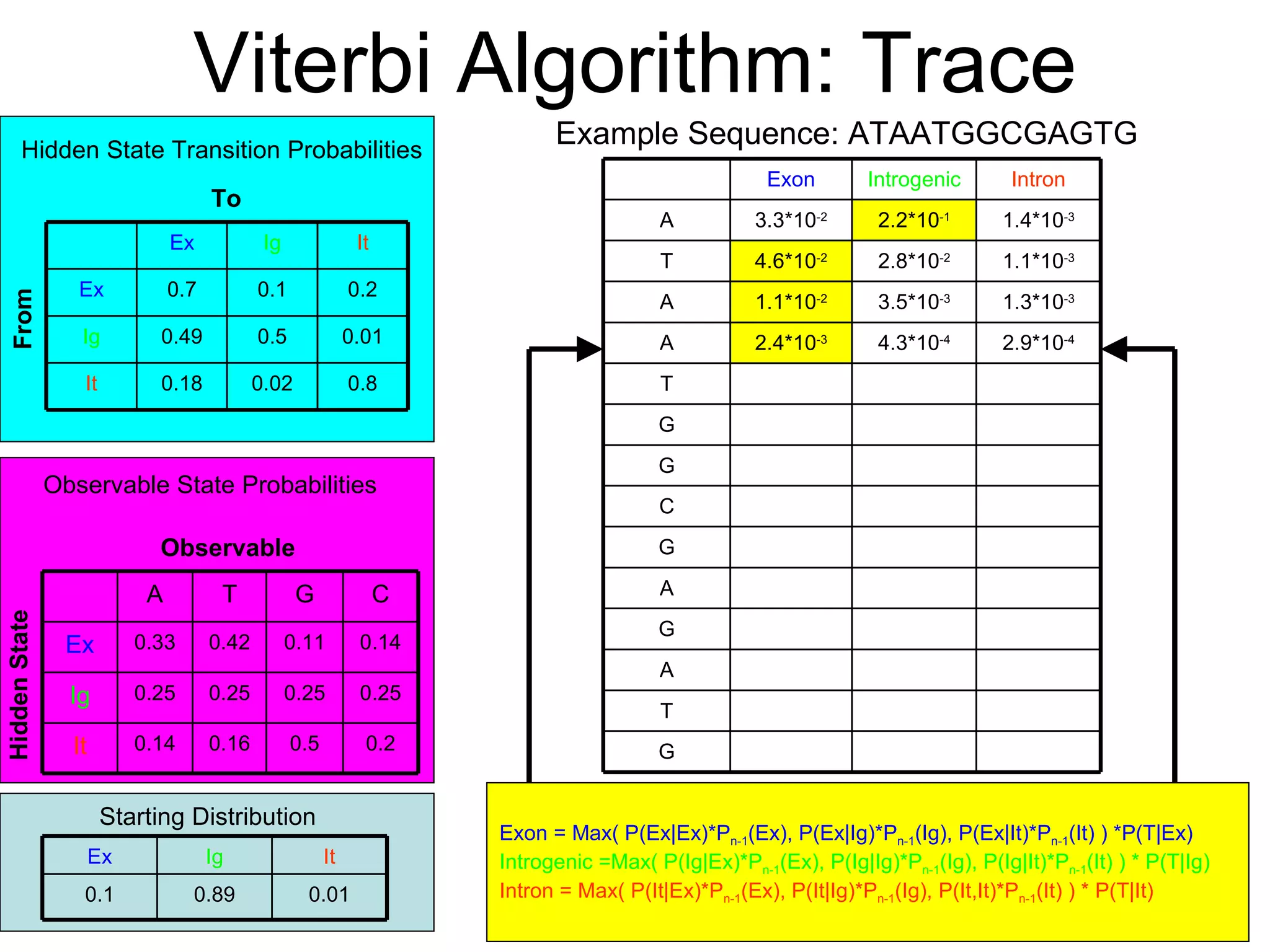
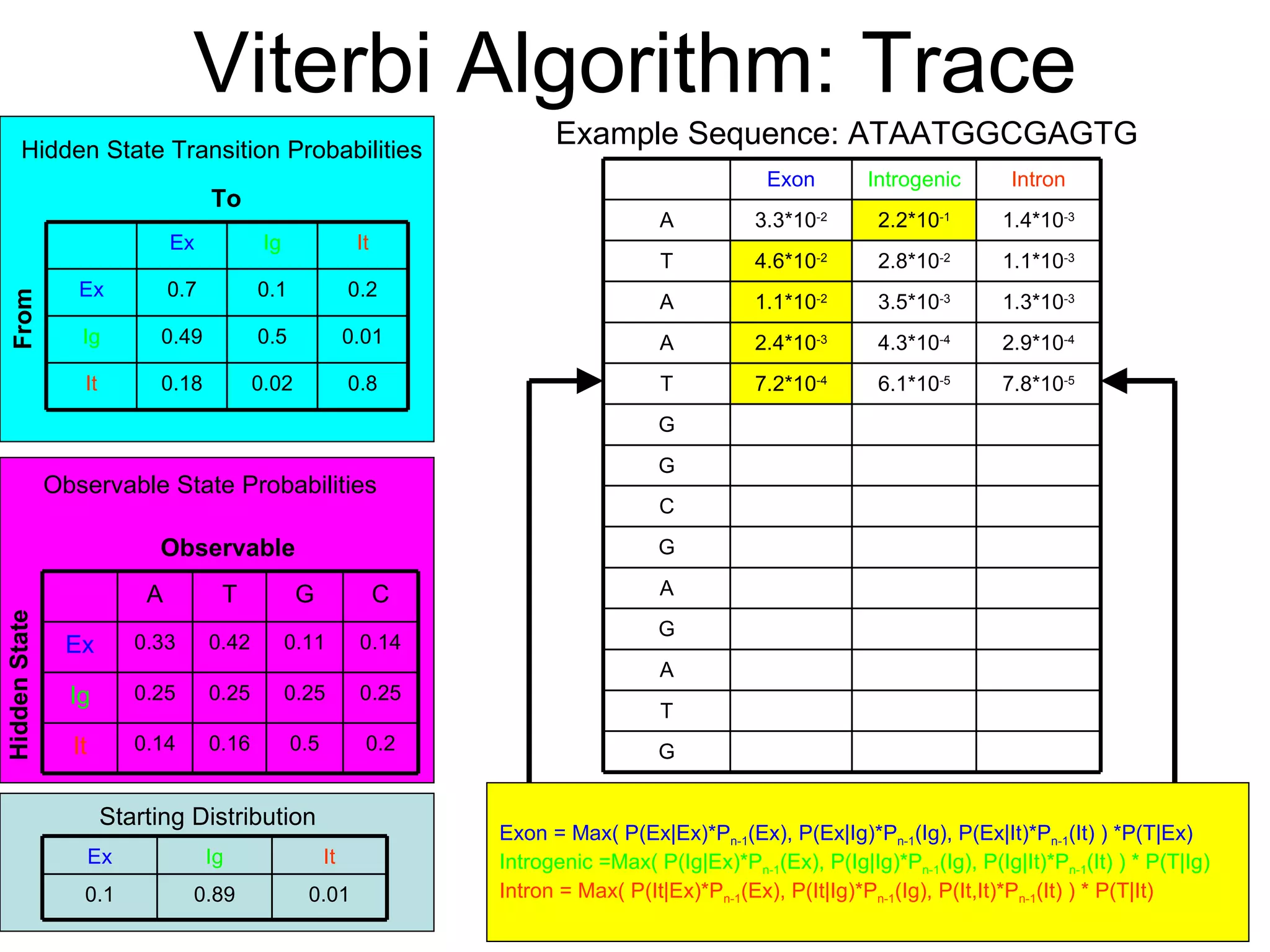
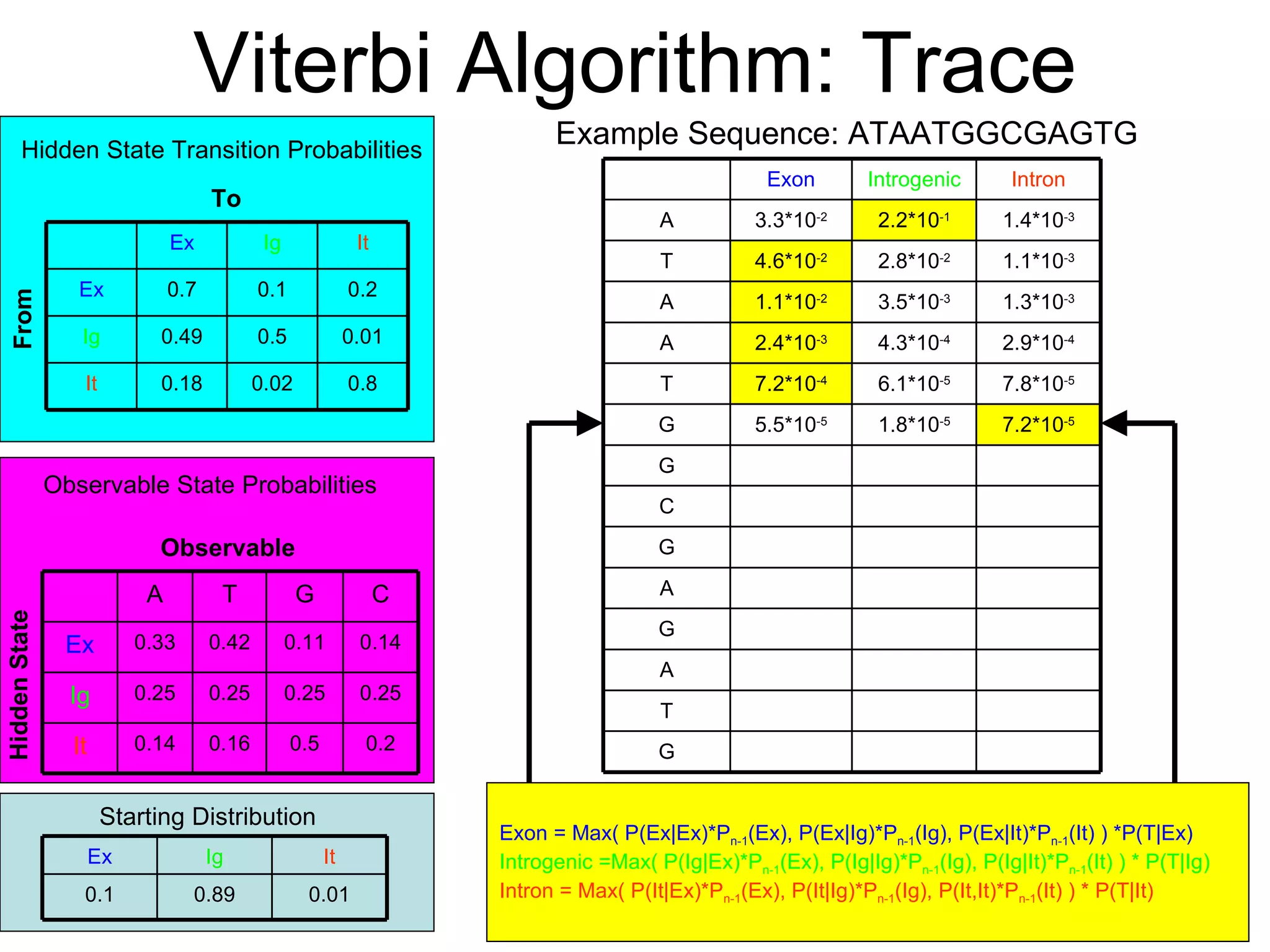
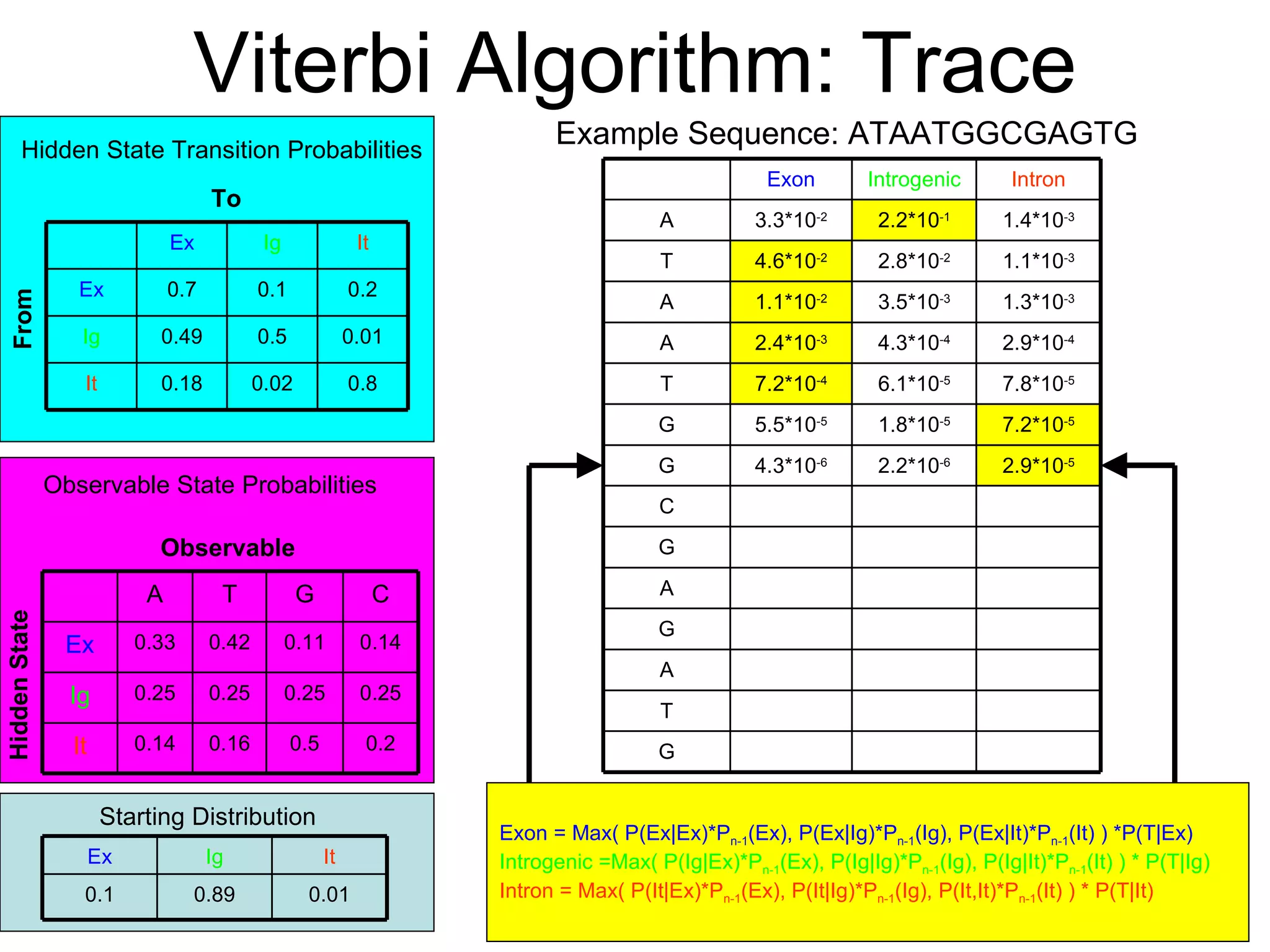
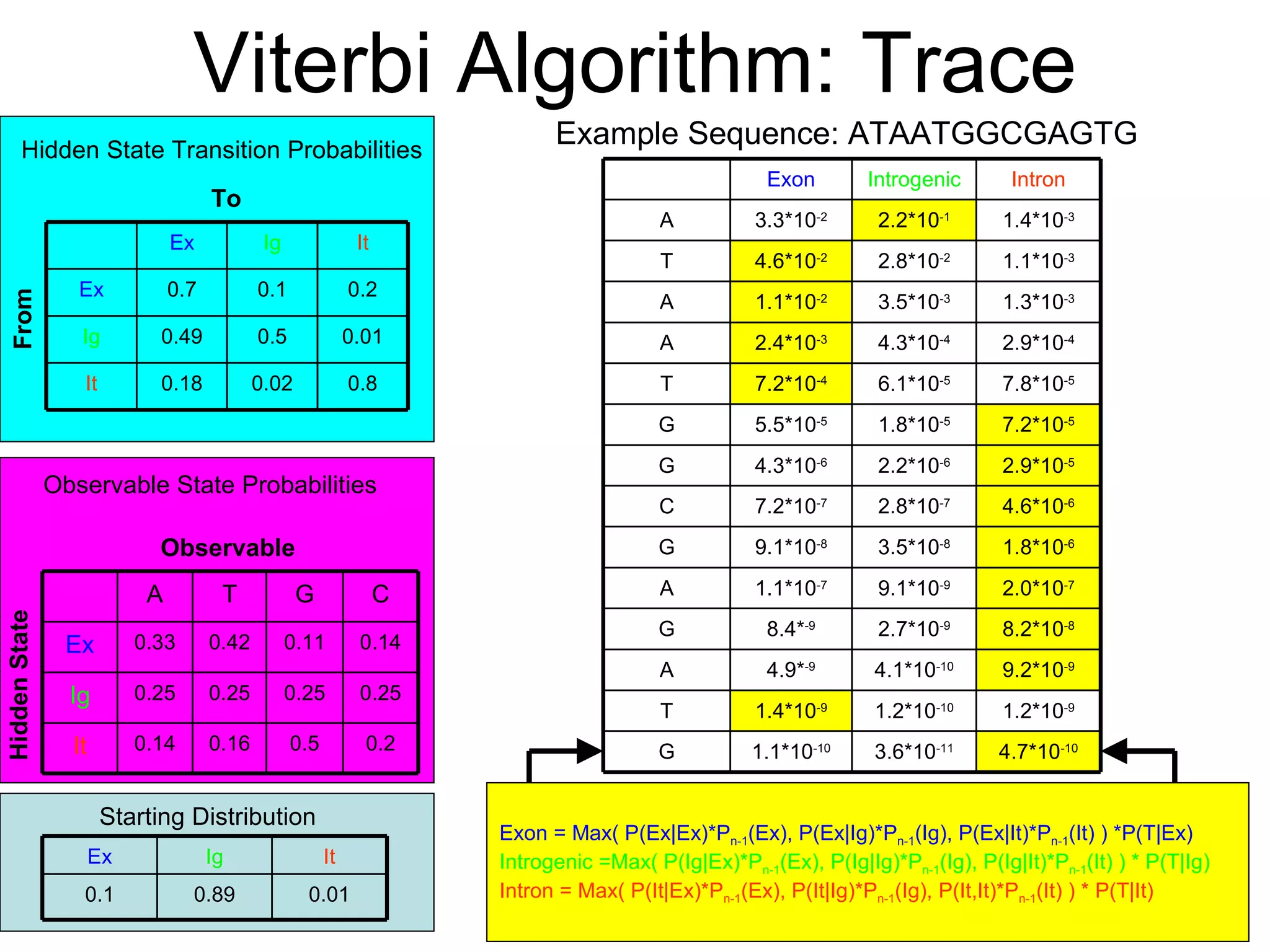
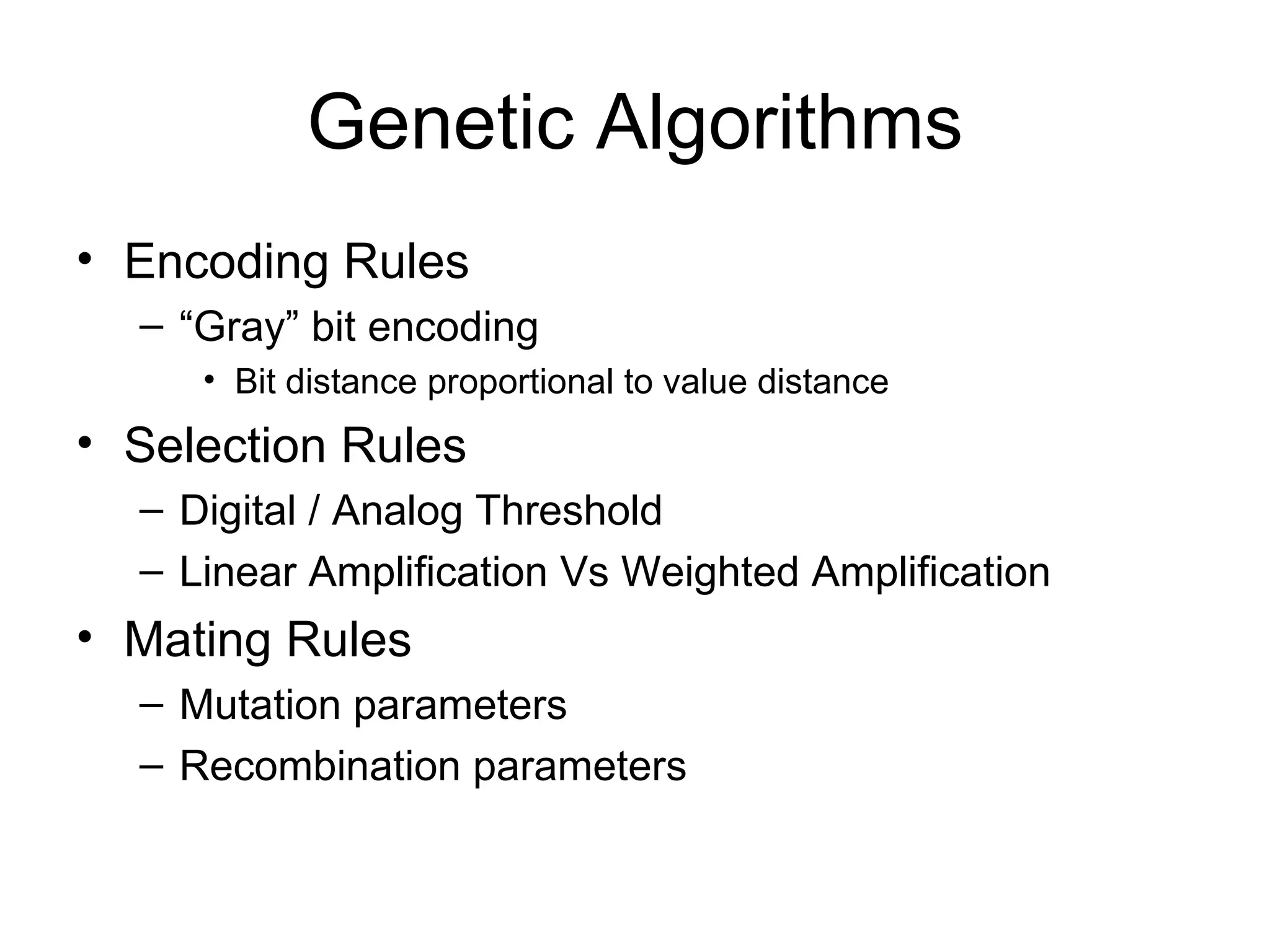
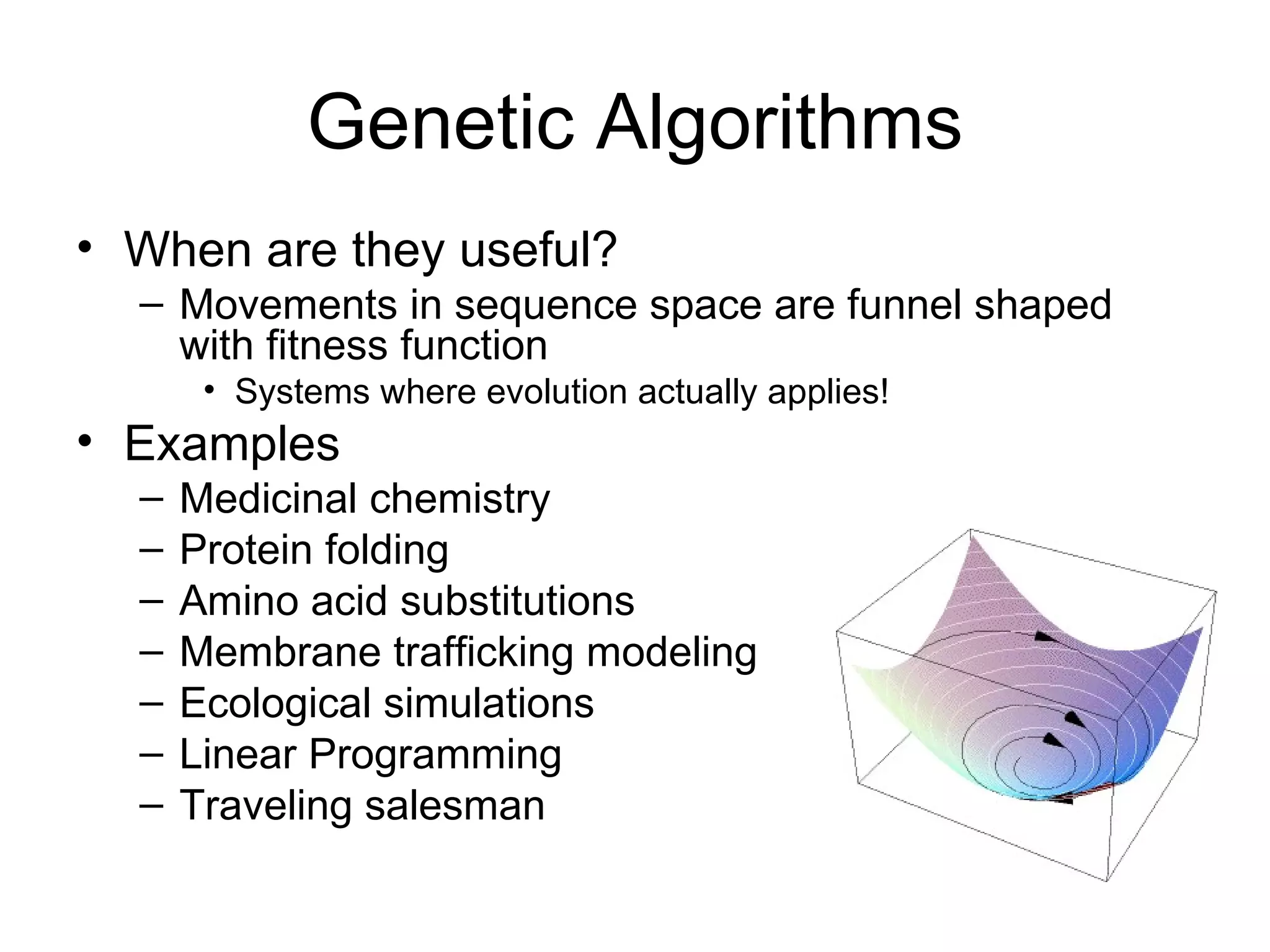
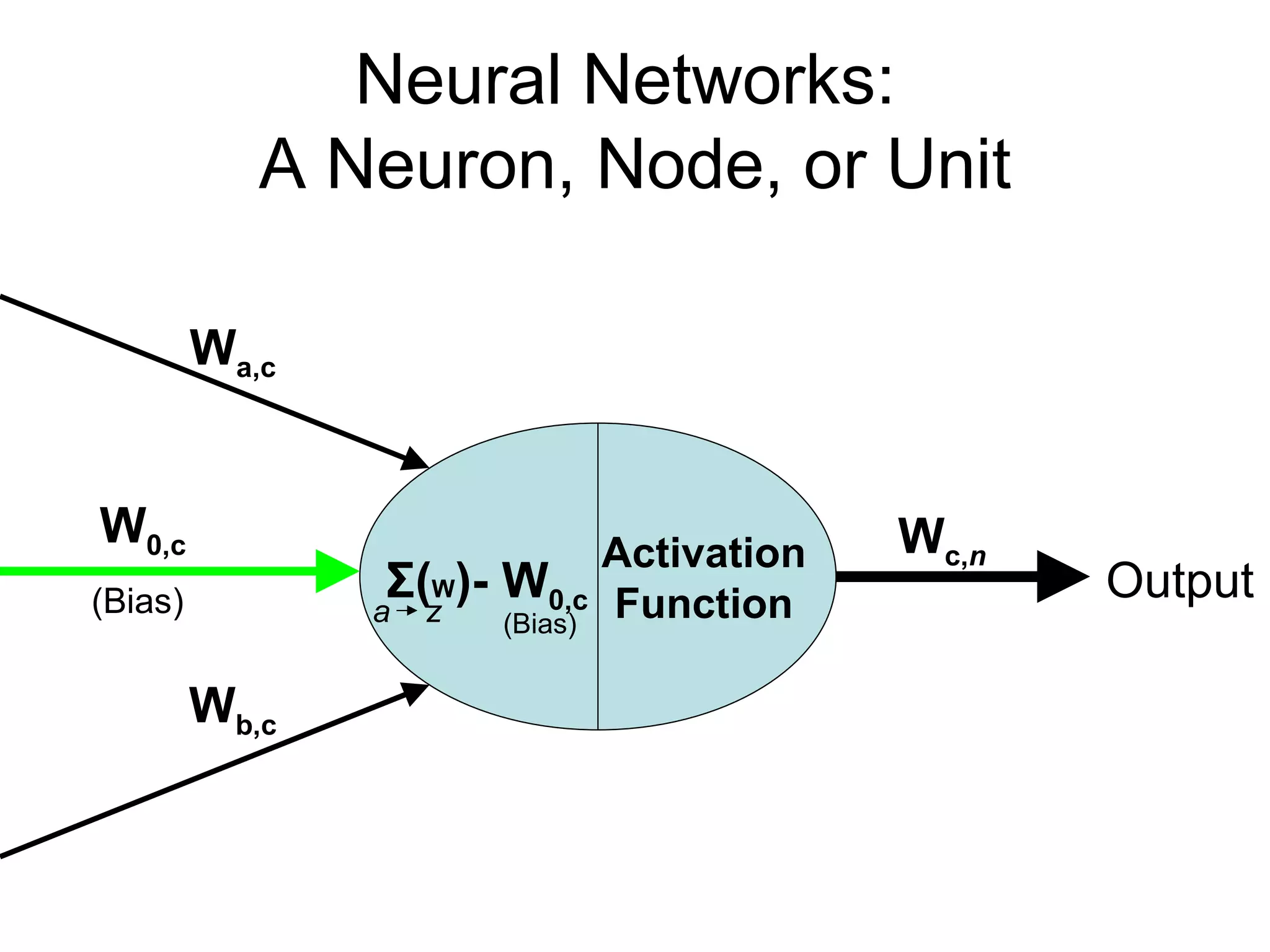
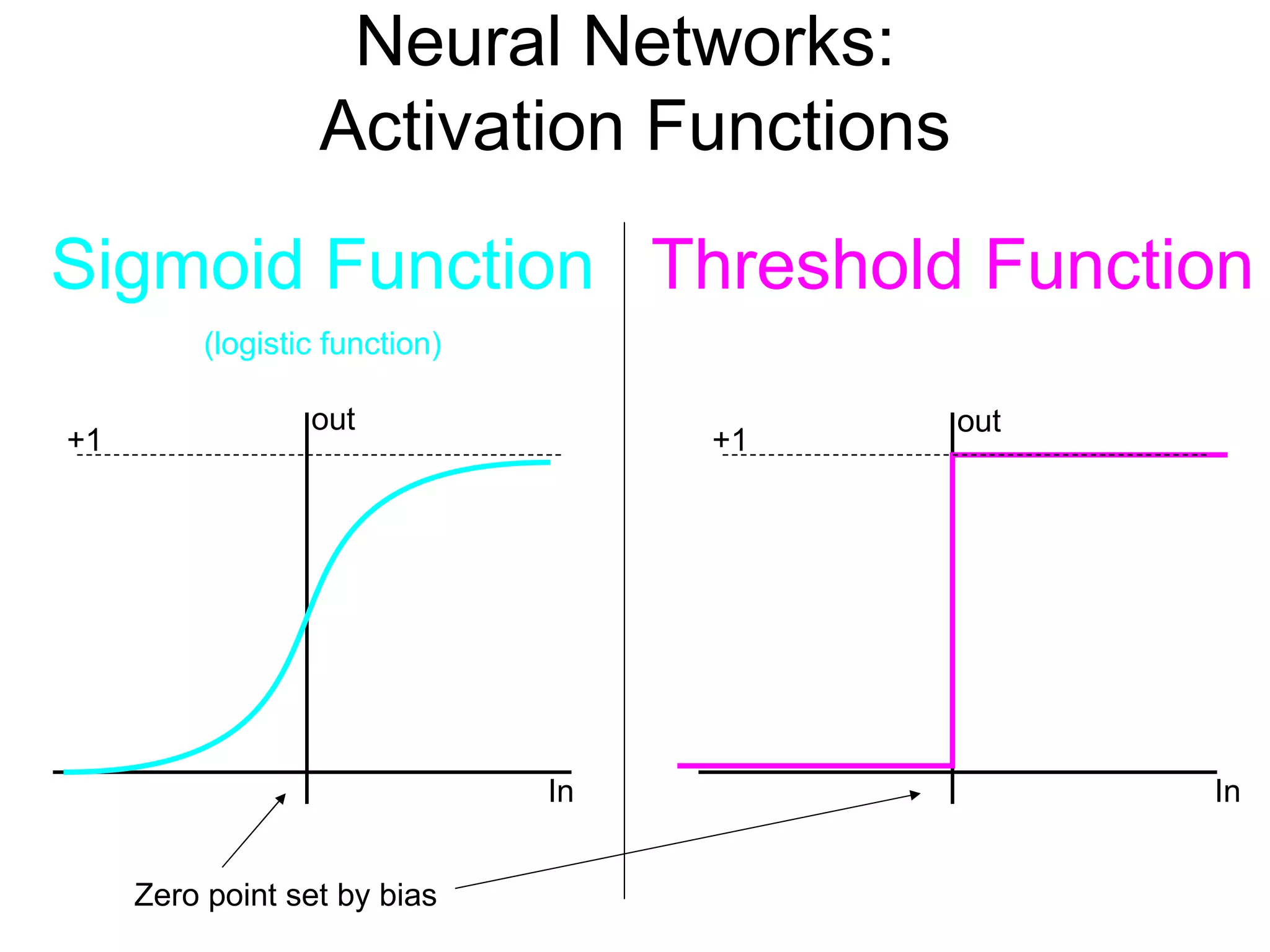
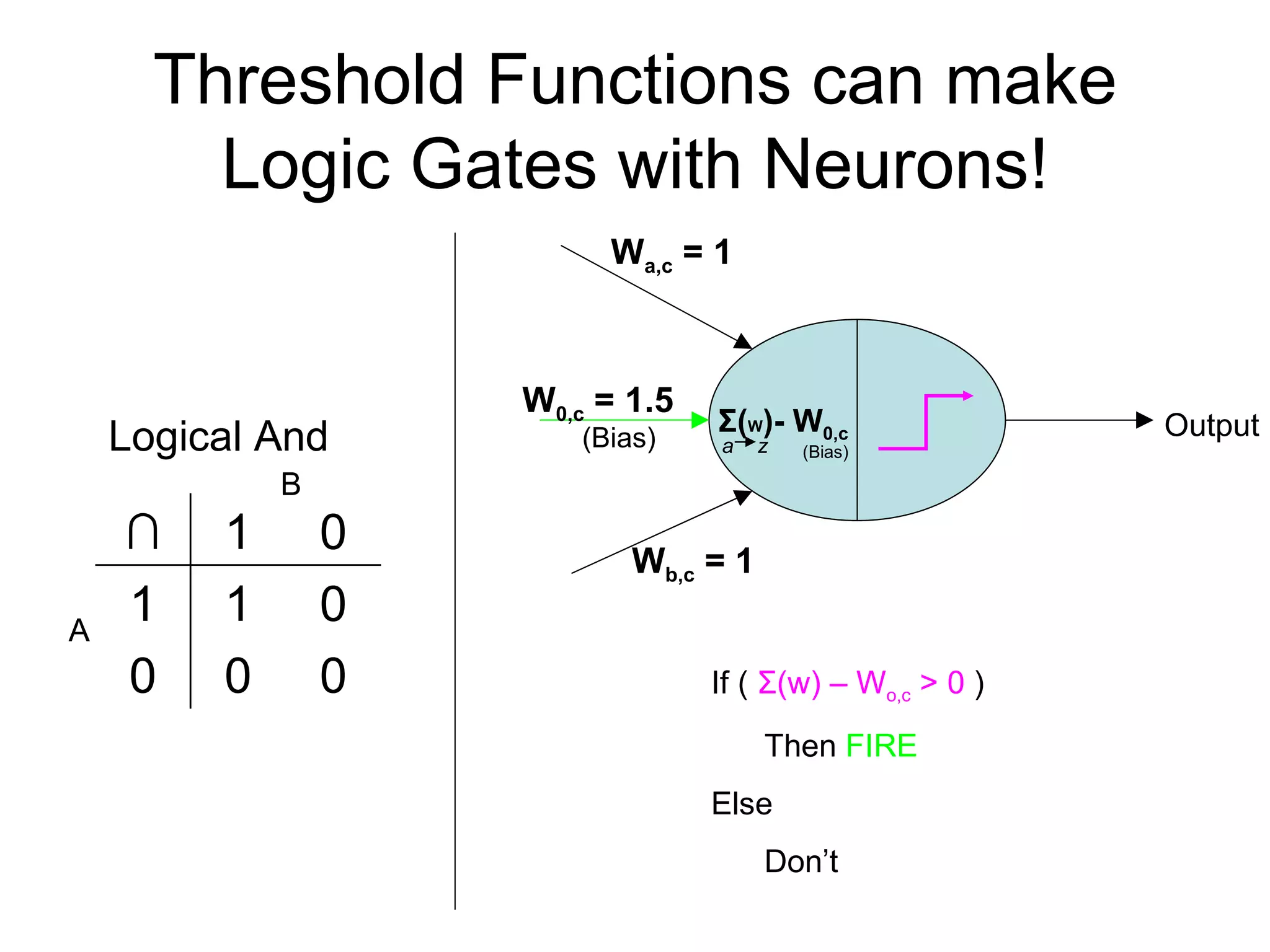
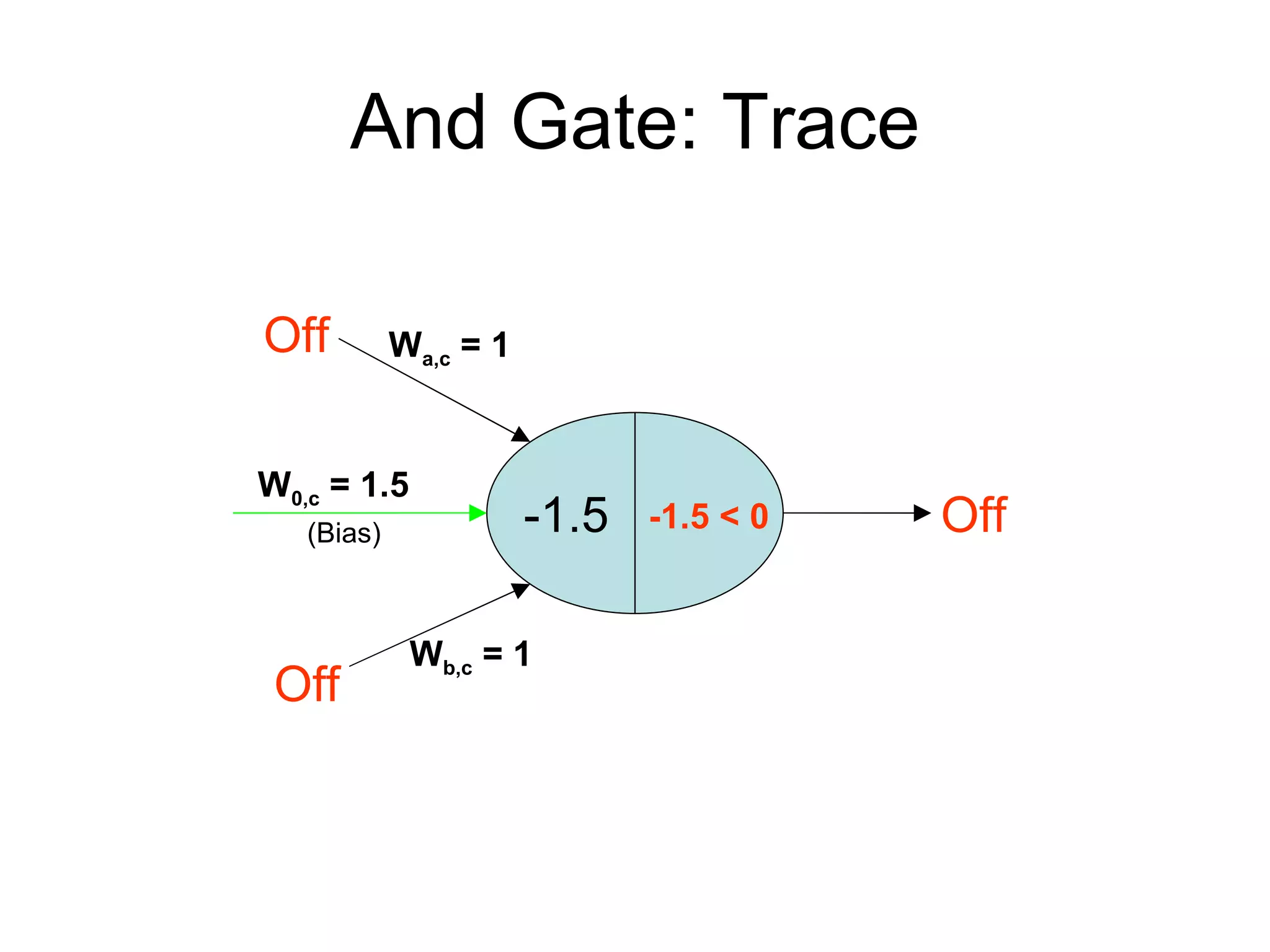
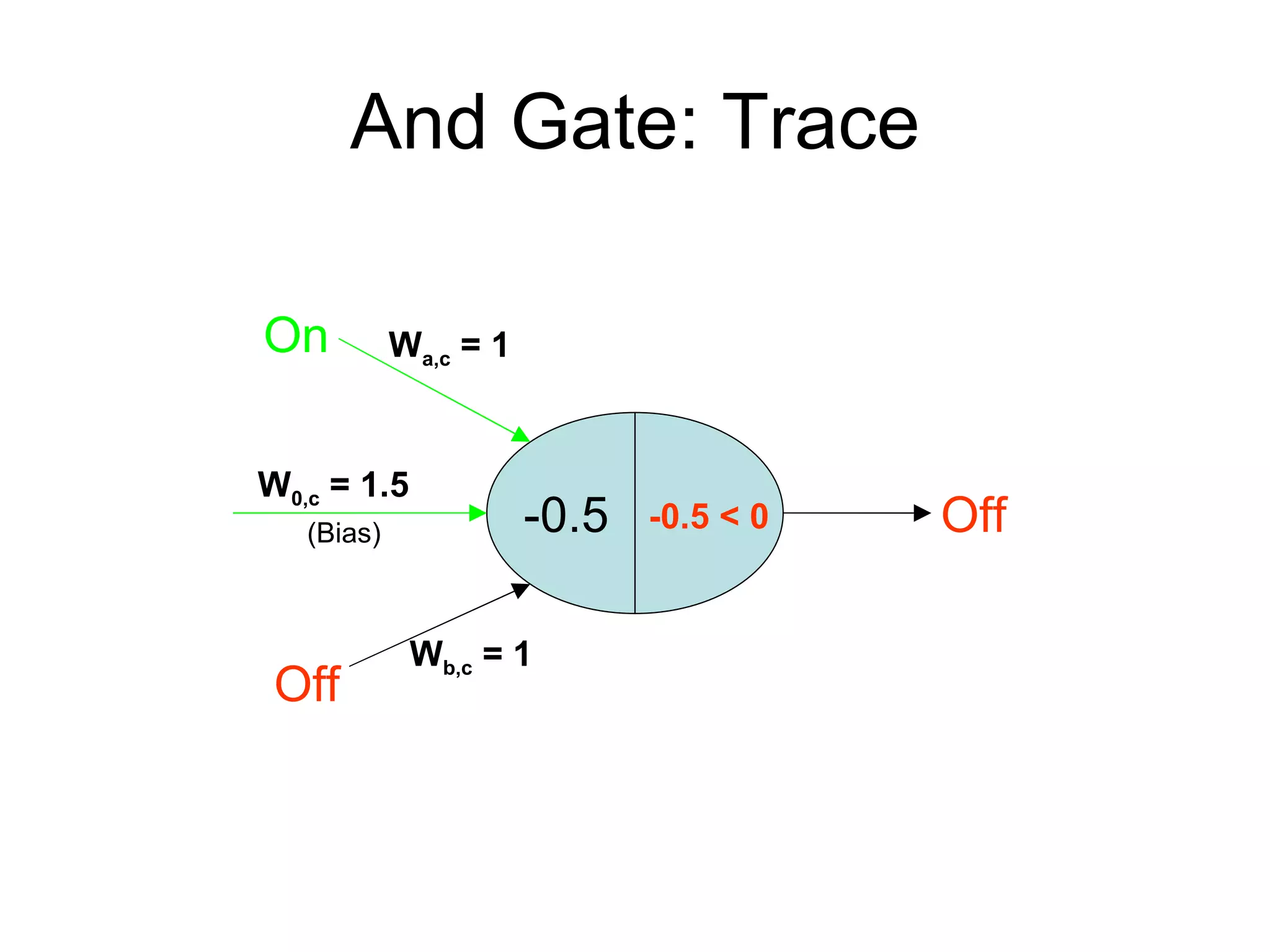
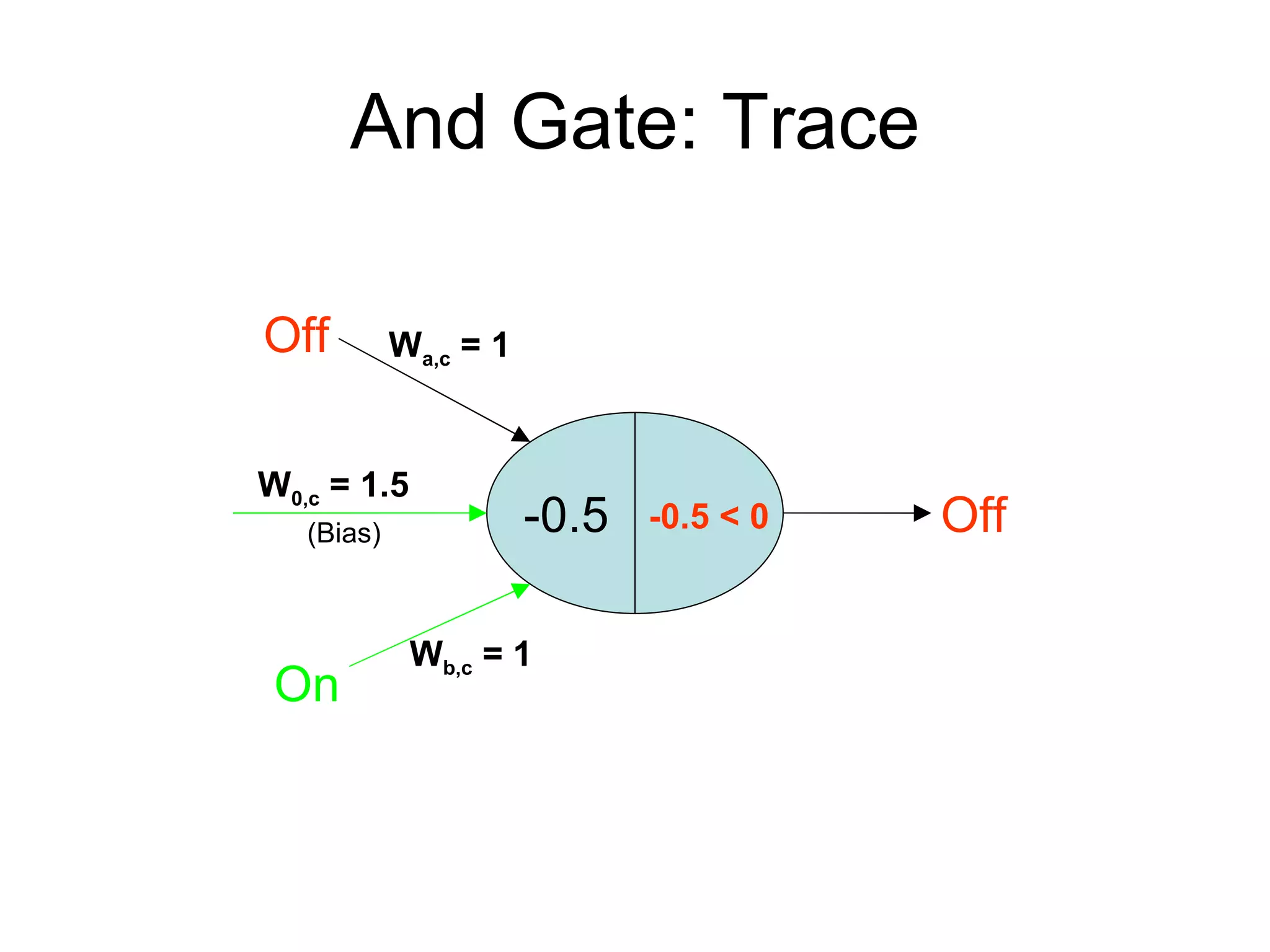
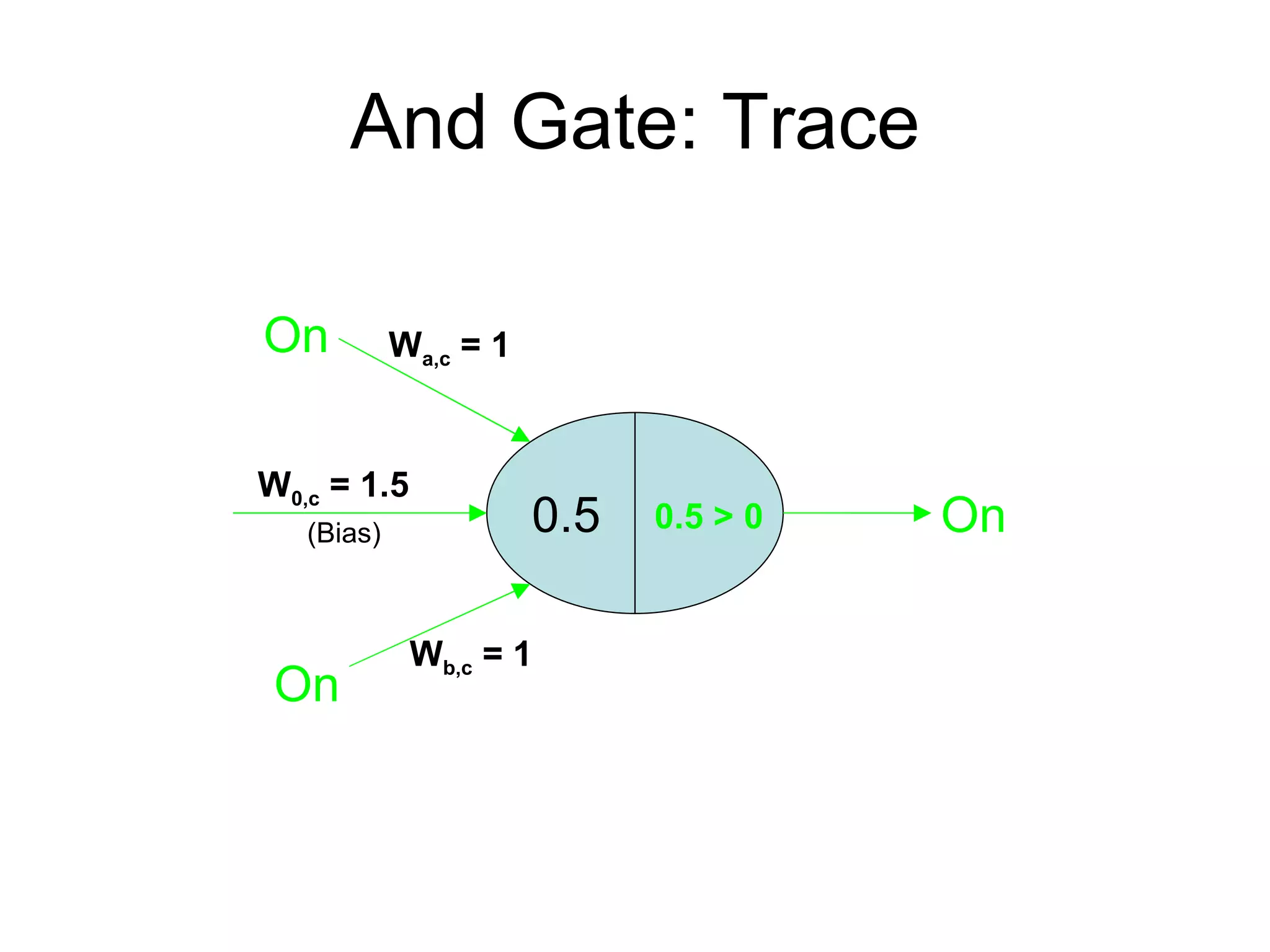
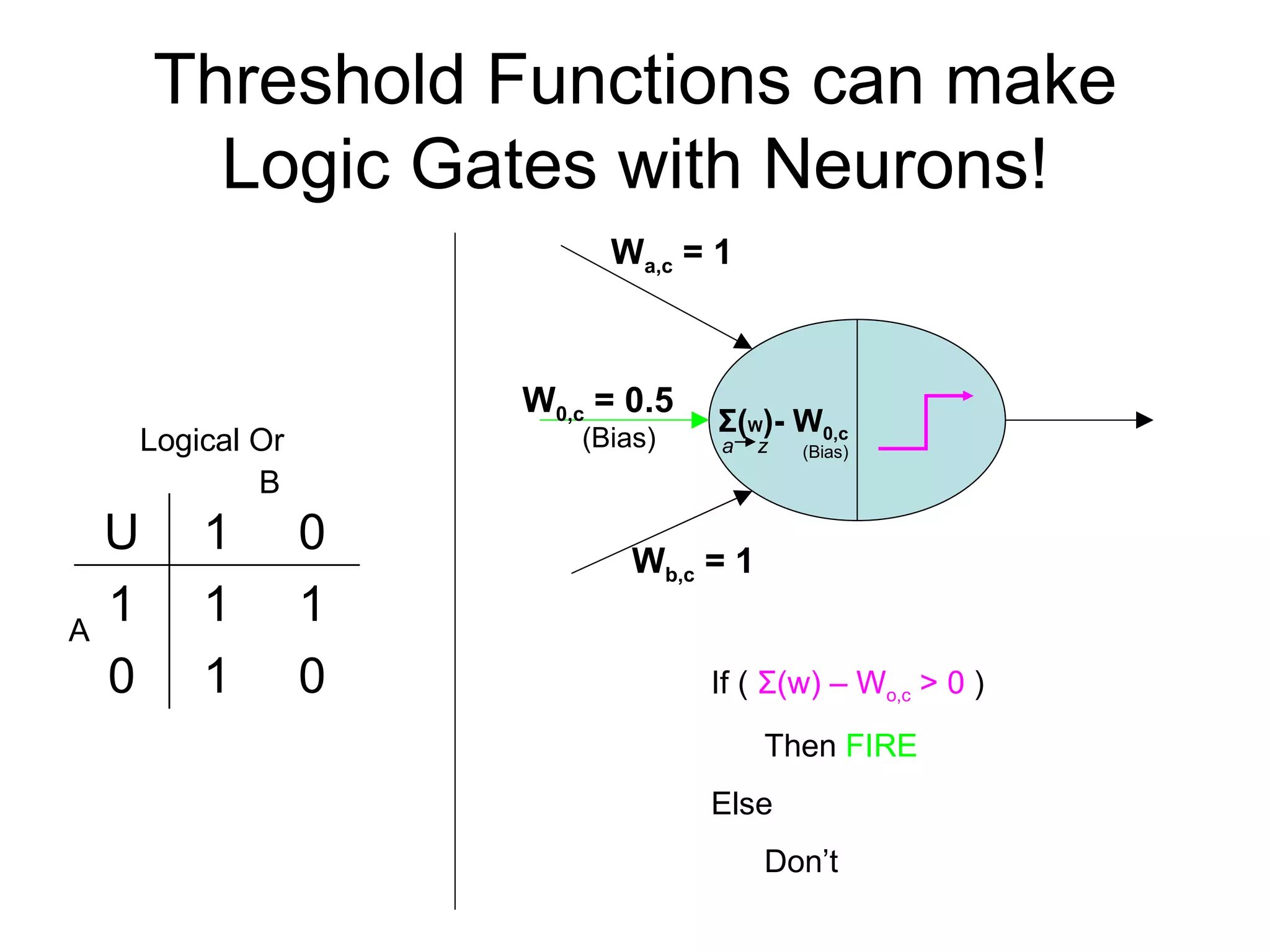
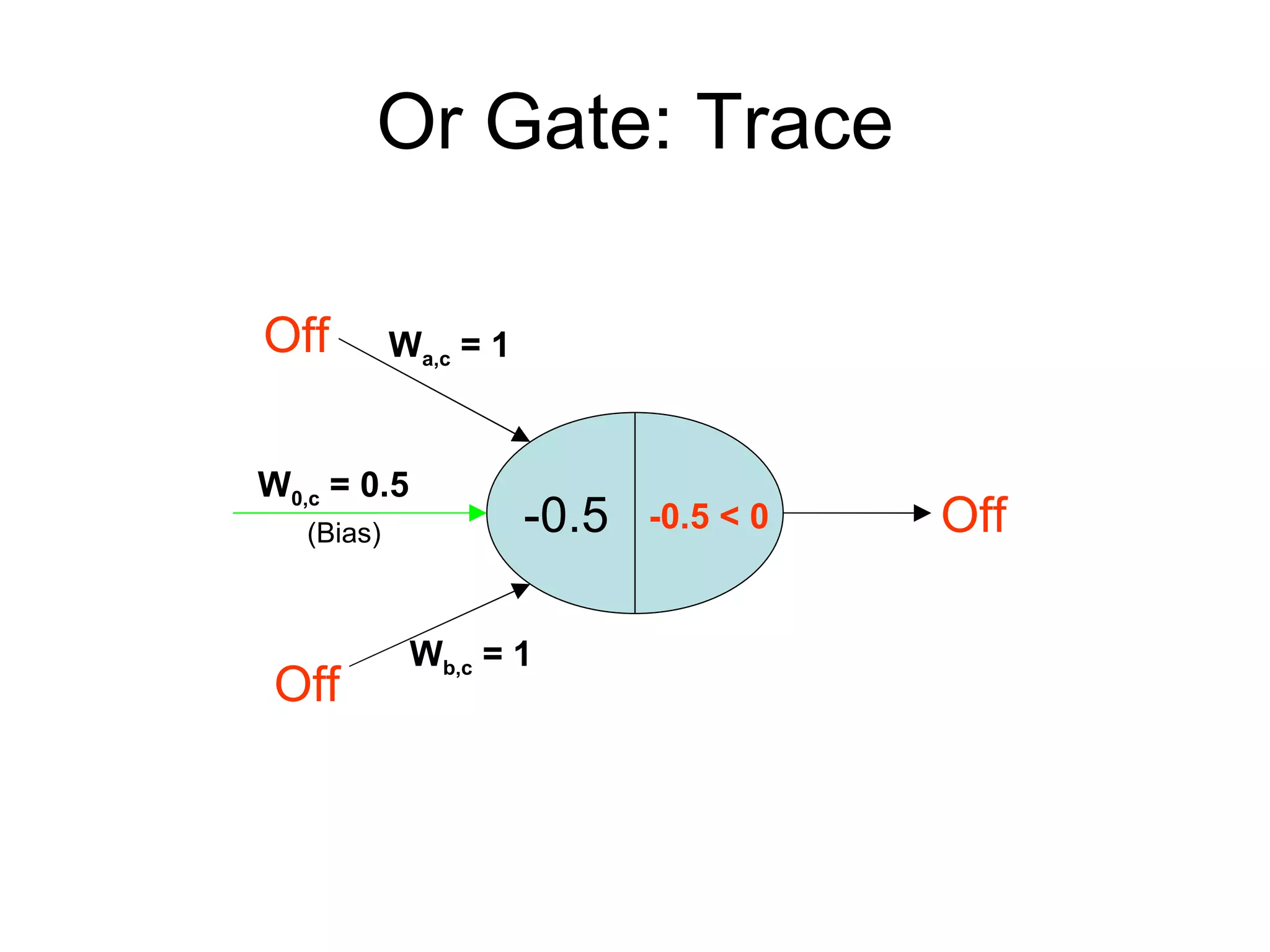
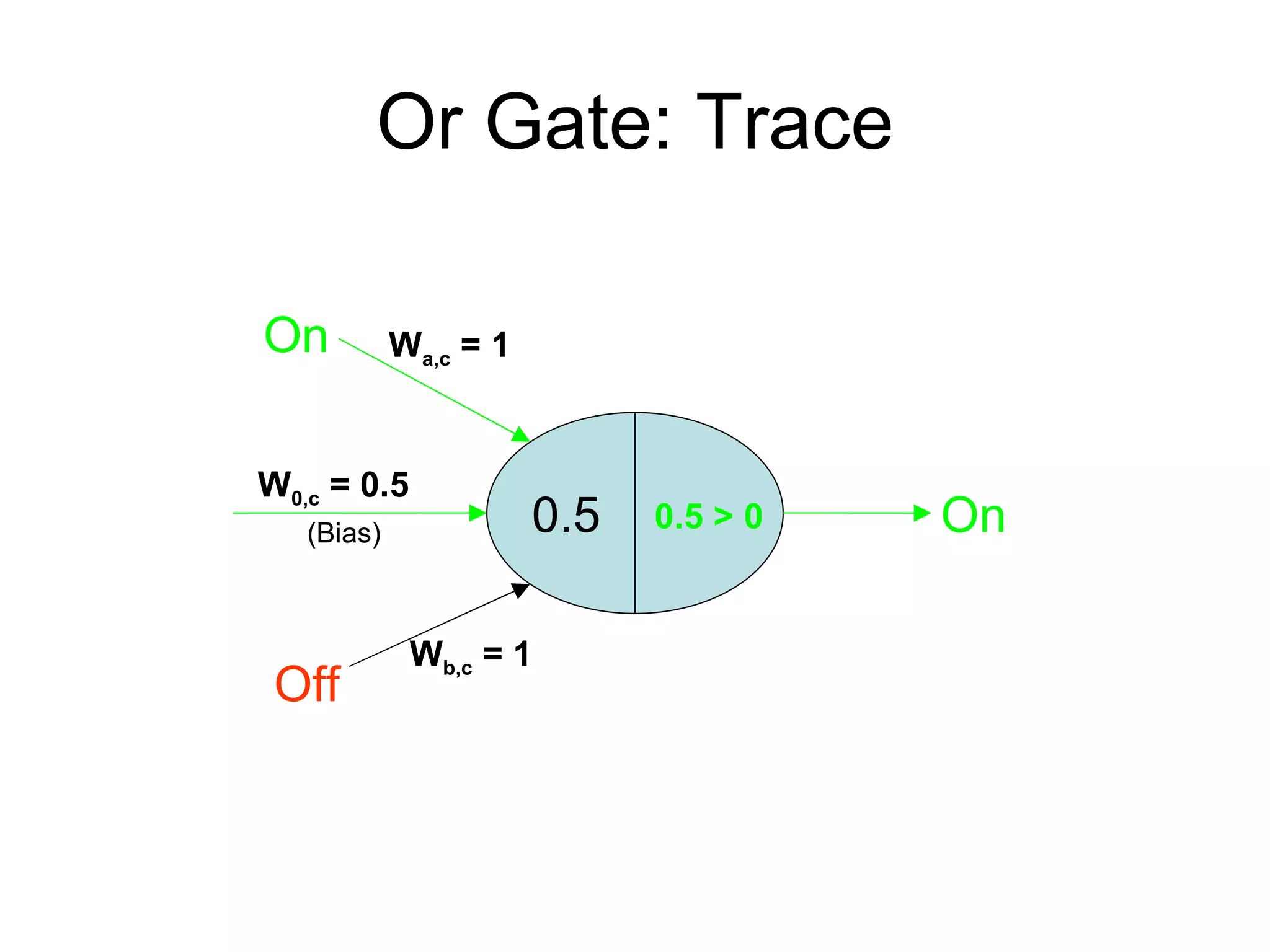
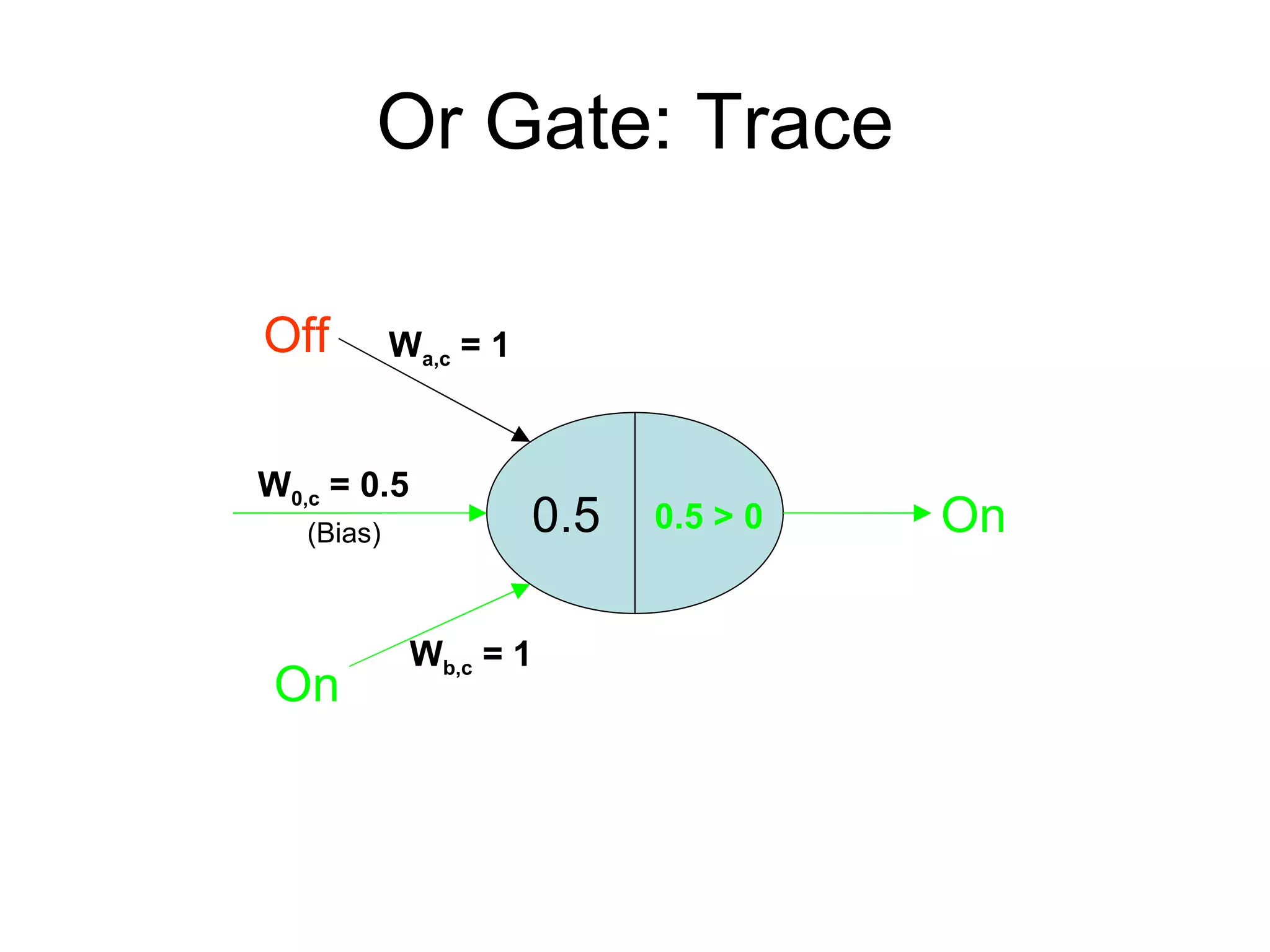
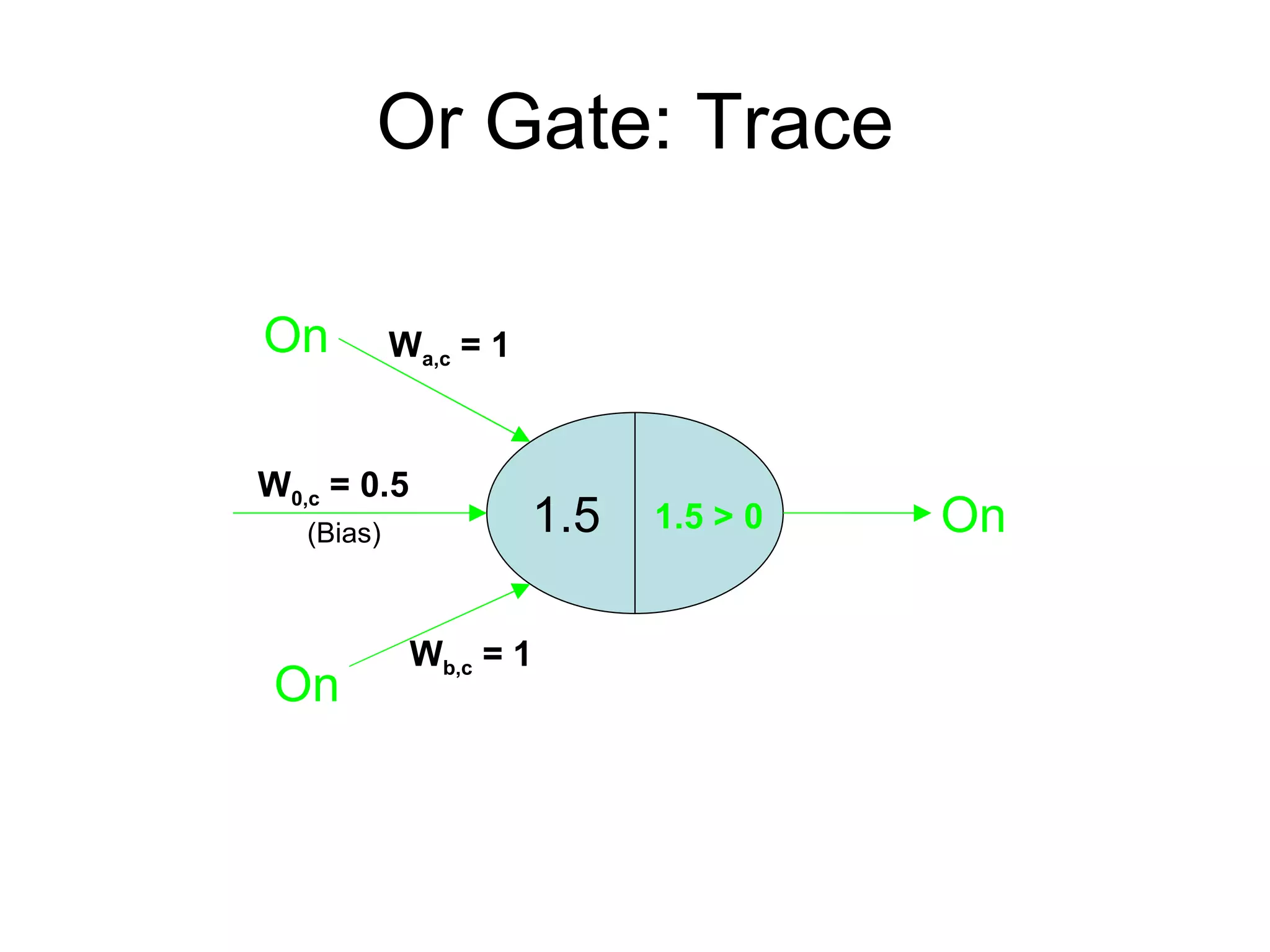
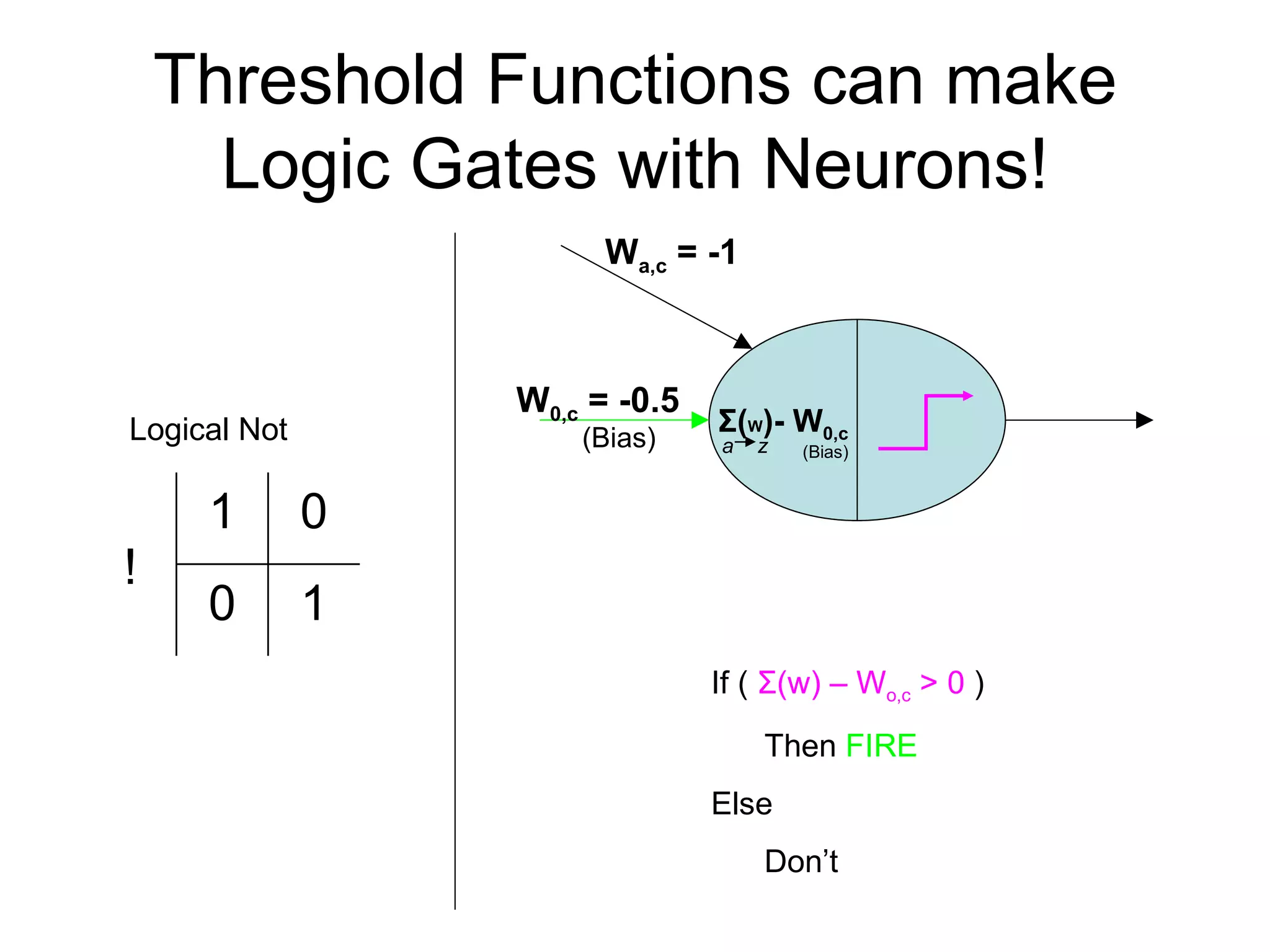
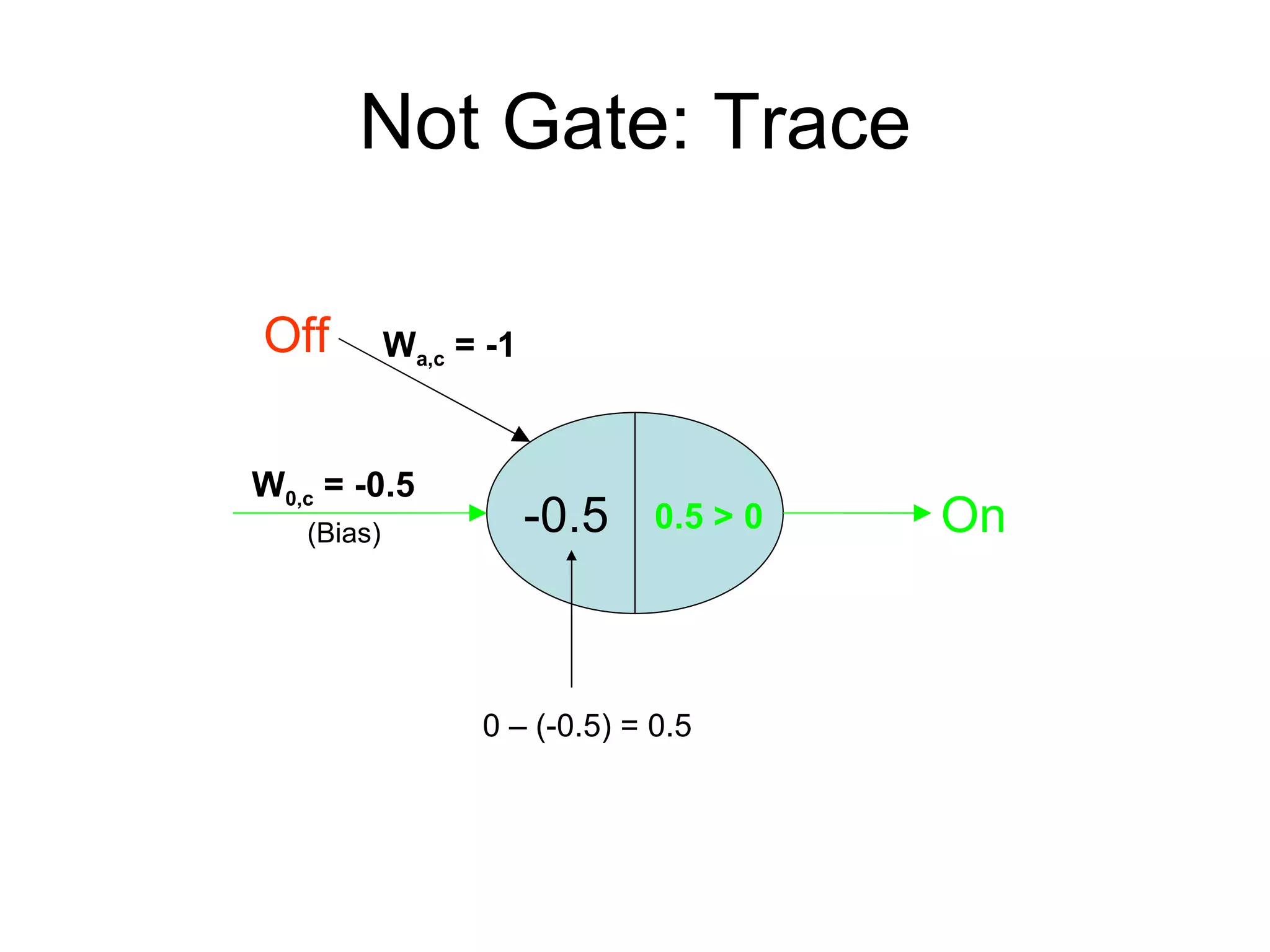
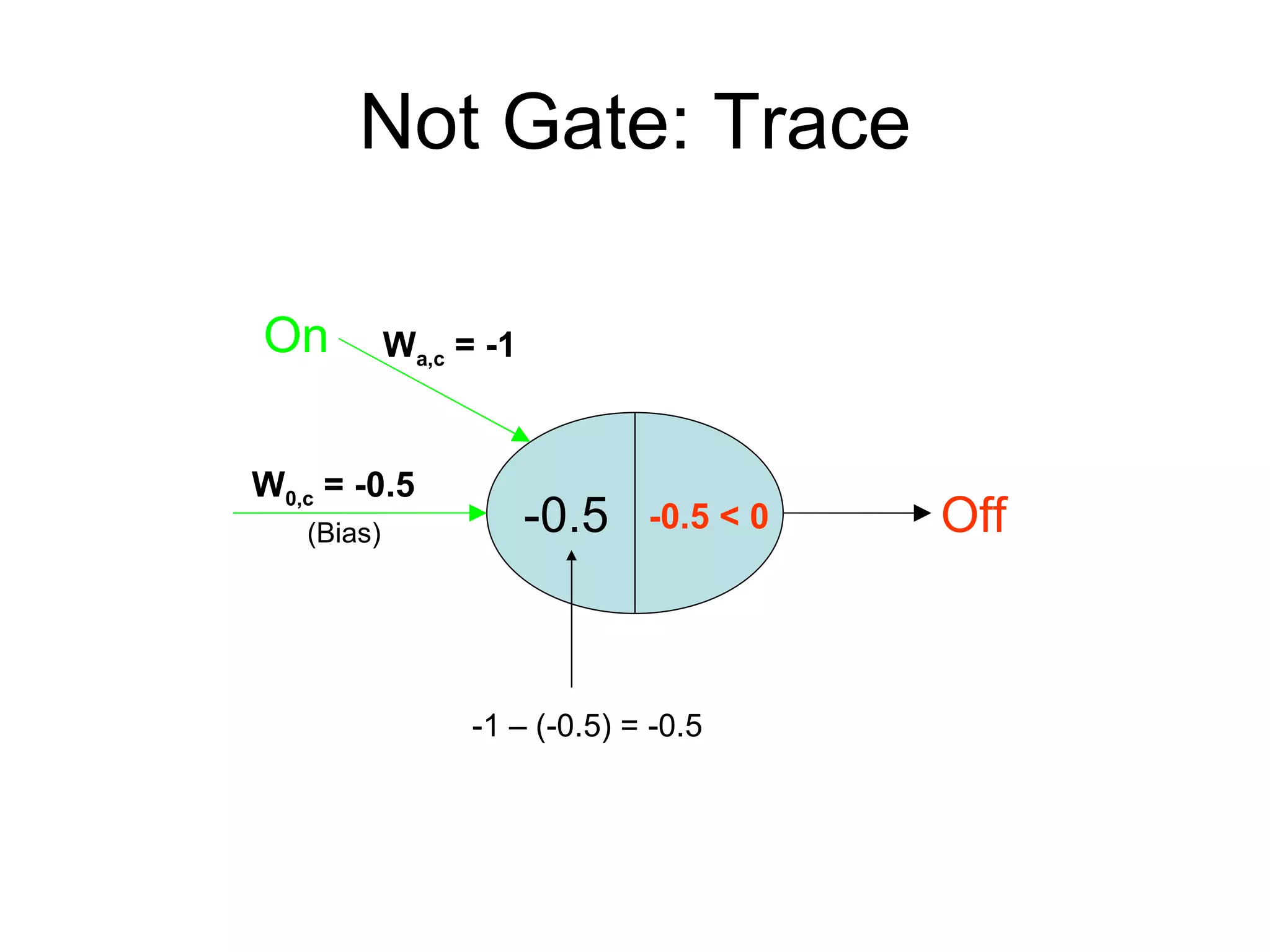
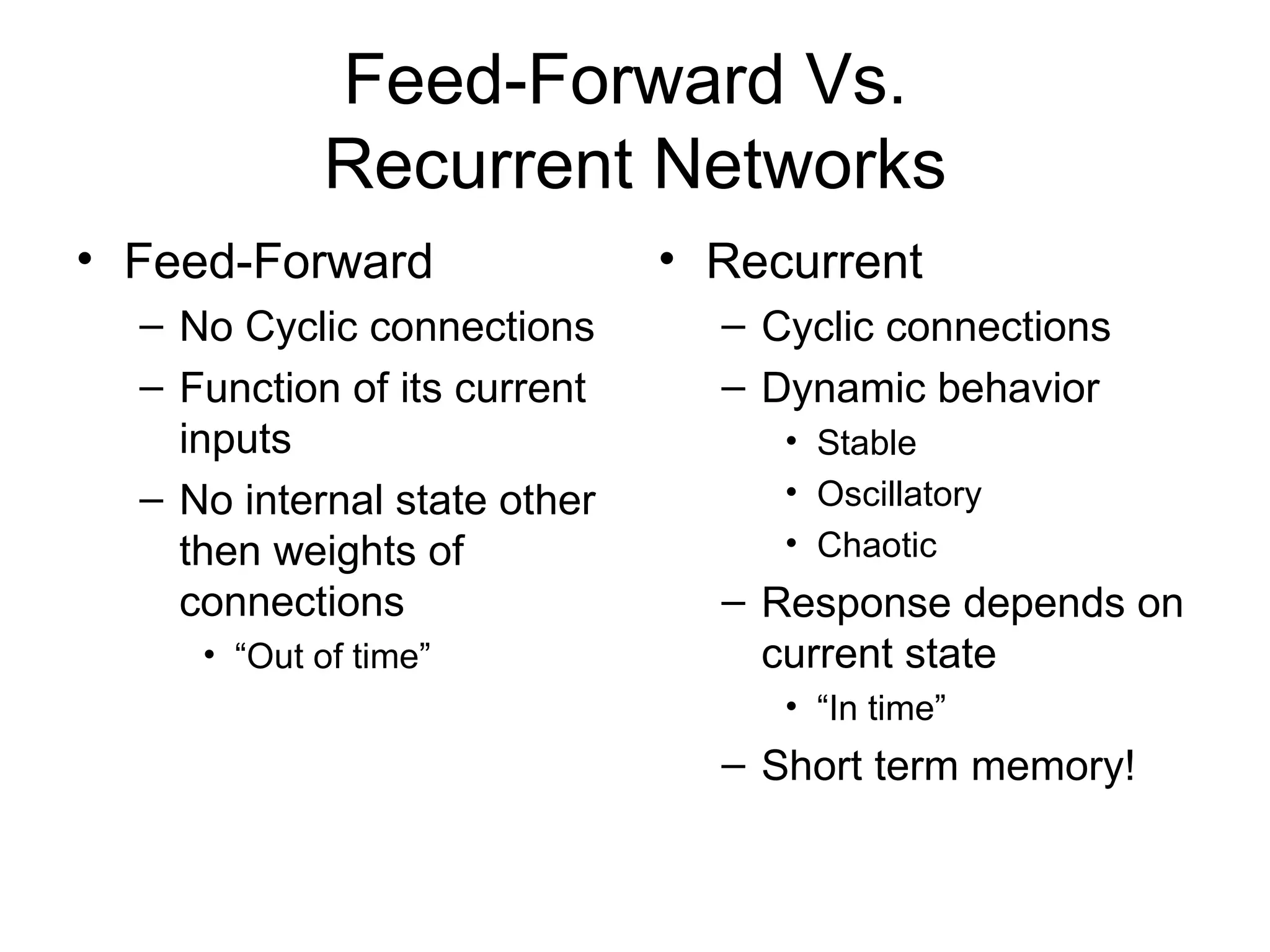
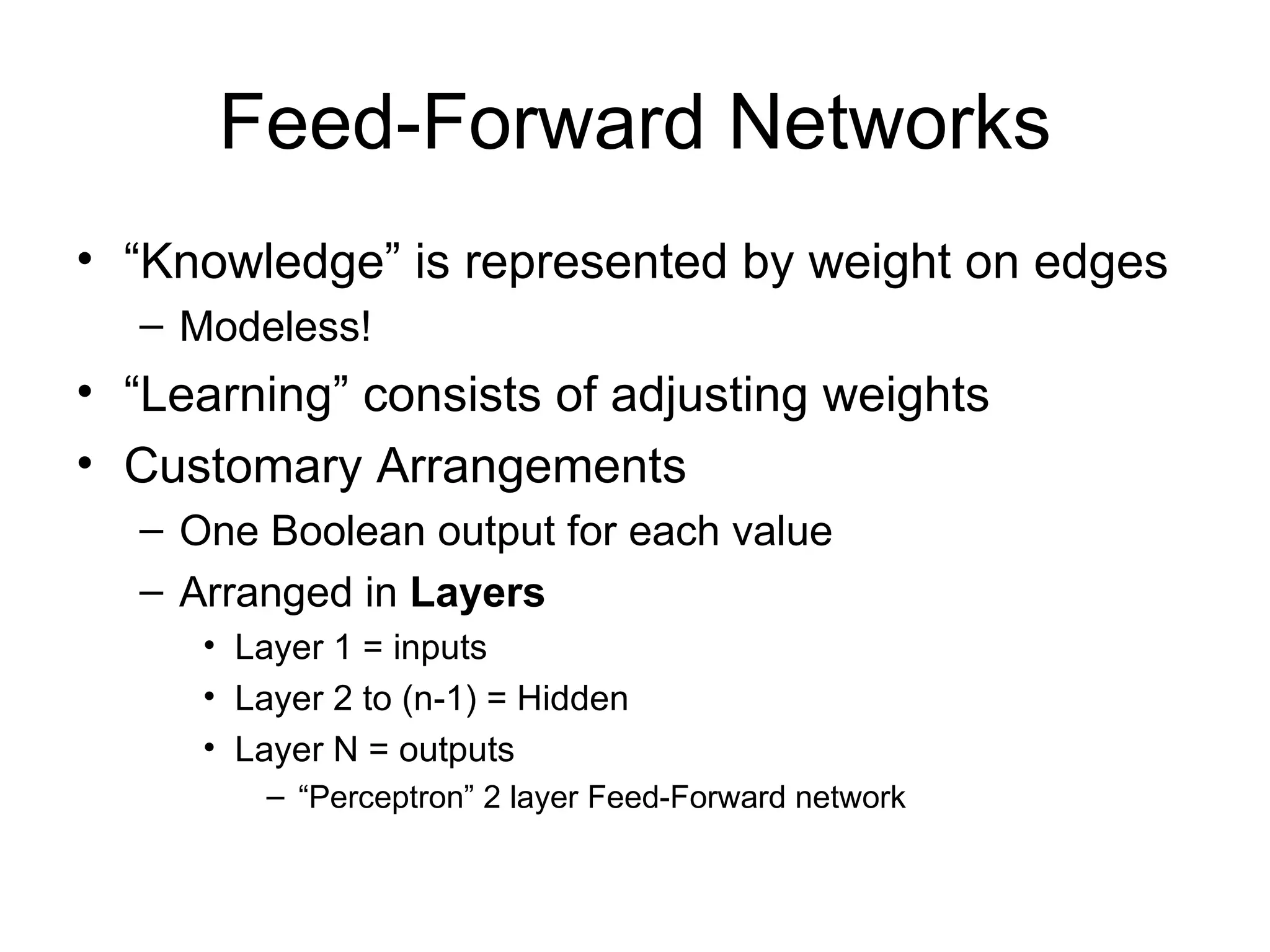
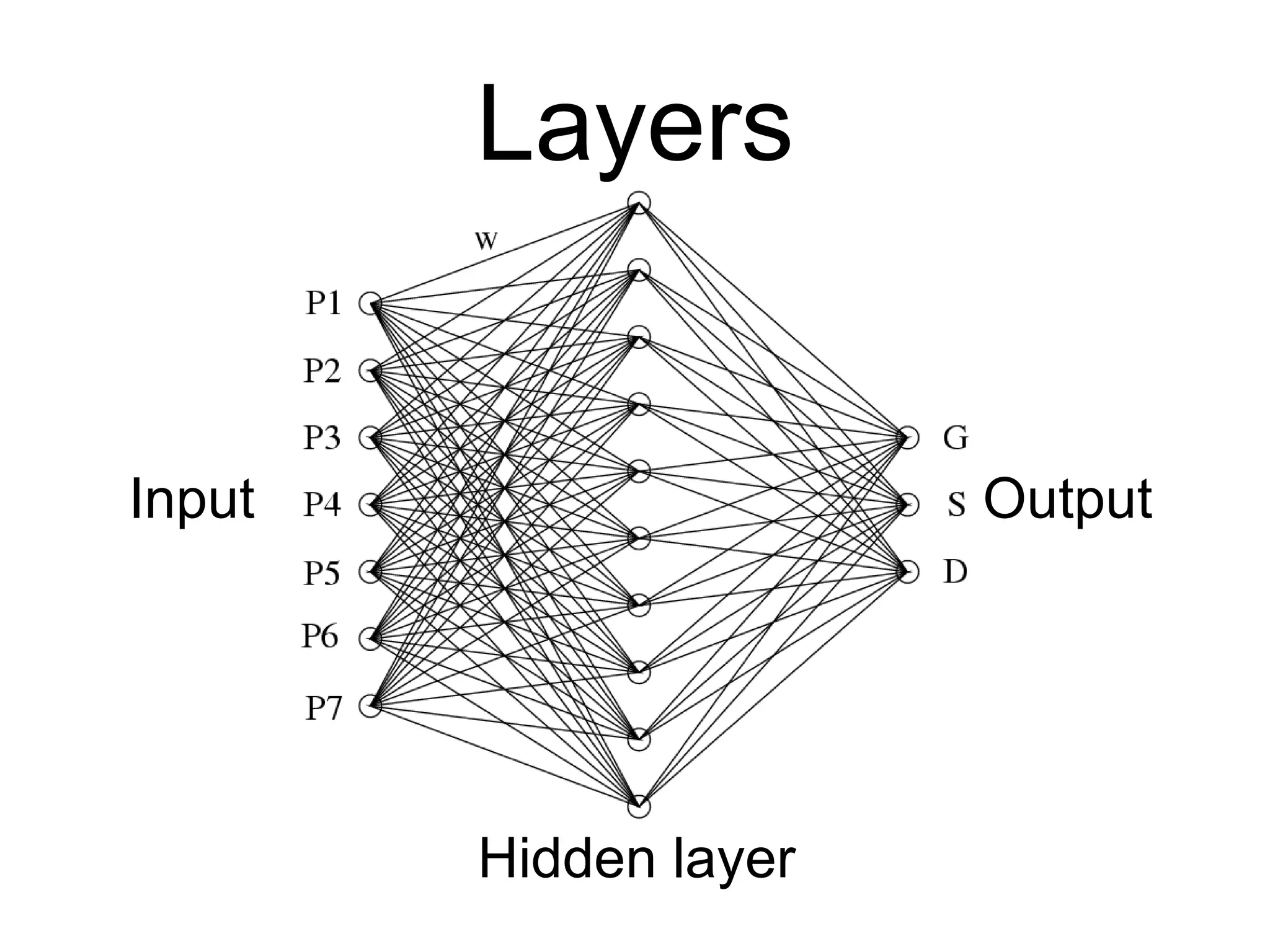
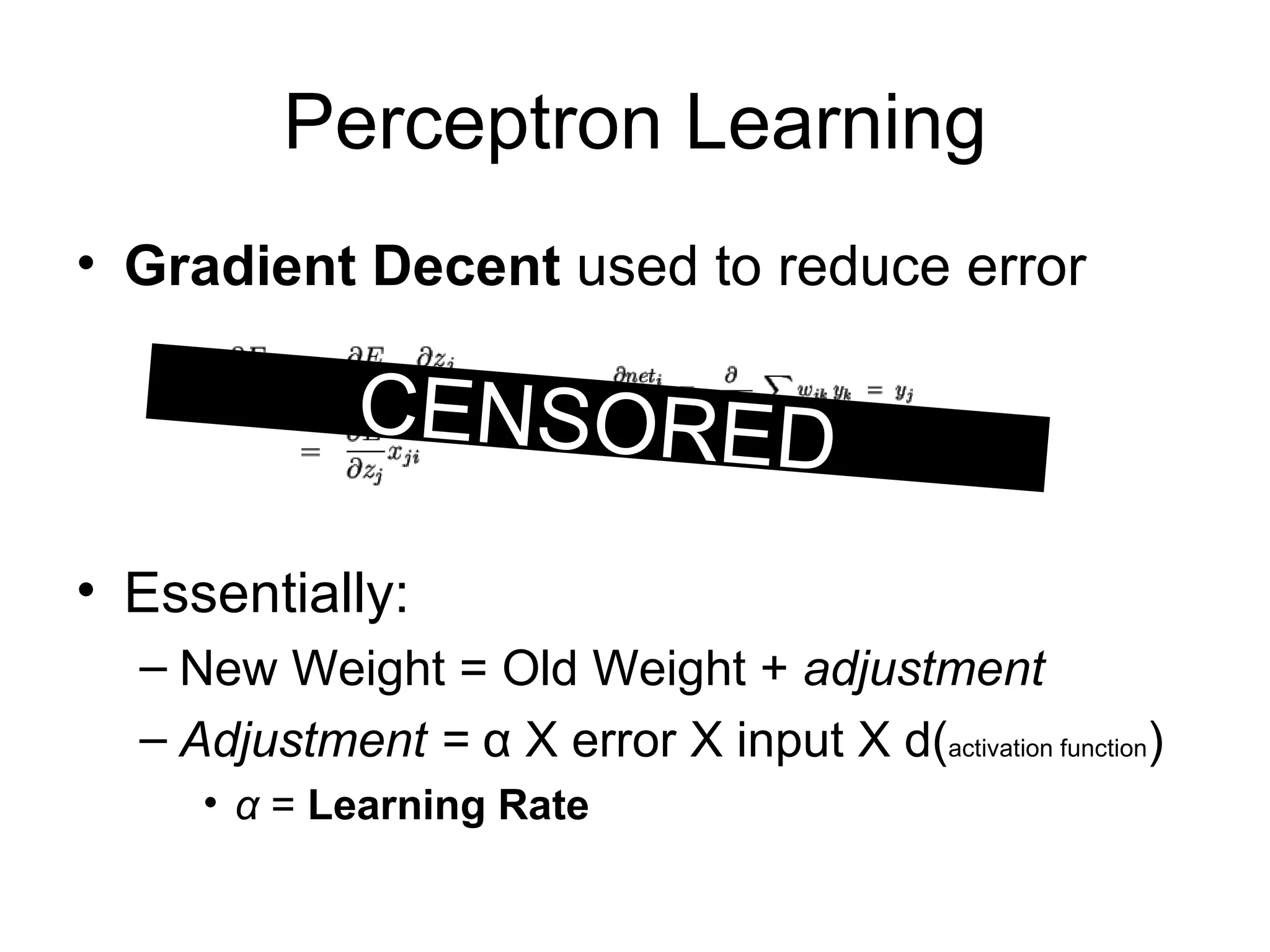
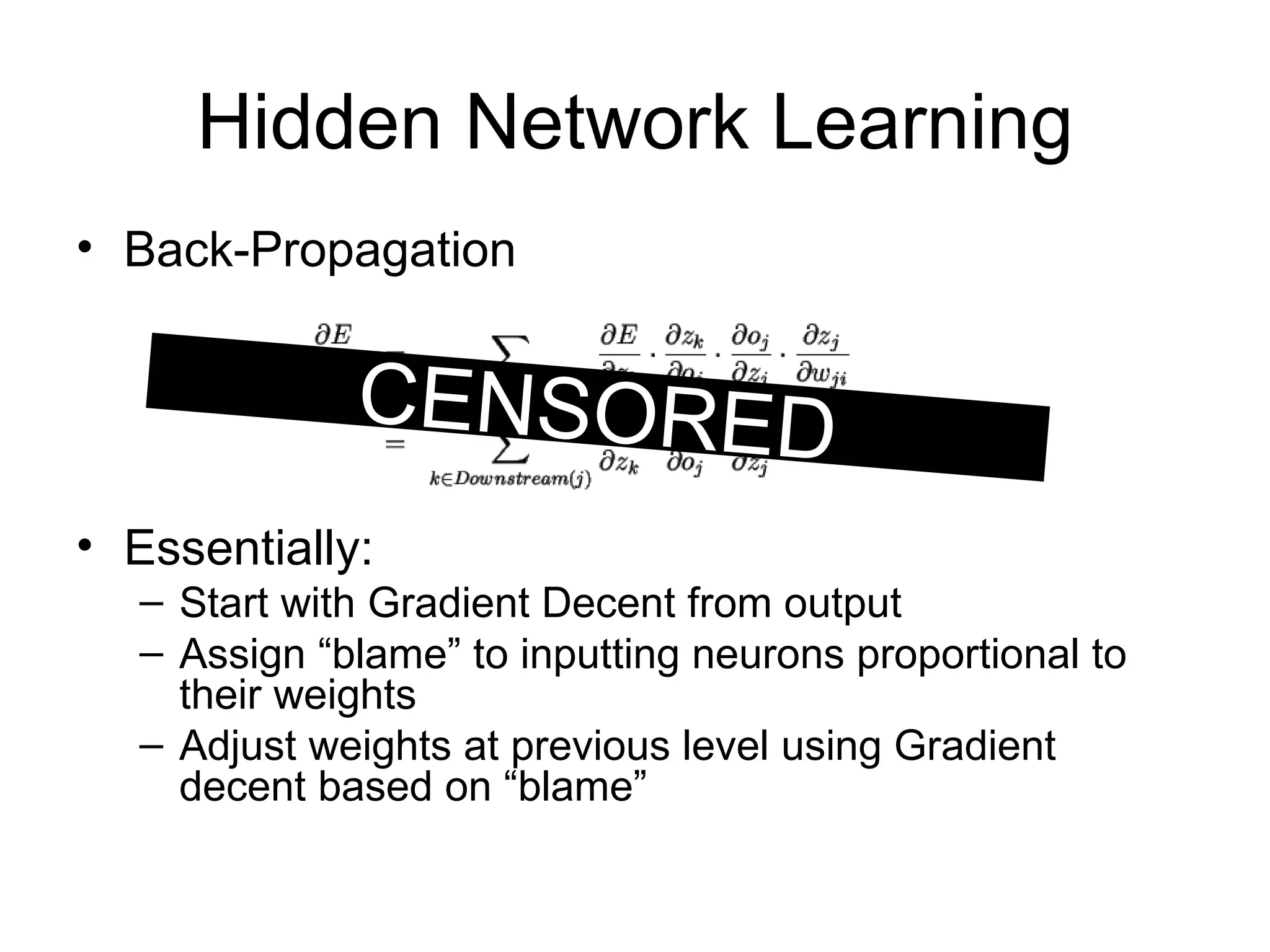
Brian M. Frezza's document explores learning algorithms in artificial intelligence, defining them as algorithms that predict data behavior based on past performance. It discusses various models such as Bayesian networks, hidden Markov models, genetic algorithms, and neural networks, alongside real-world applications like speech recognition and biological predictions. Additionally, it highlights the importance of training and testing data, and how these algorithms learn from data structures and patterns.