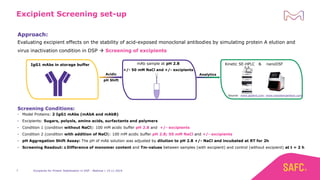
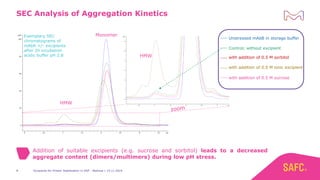
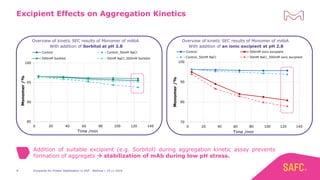
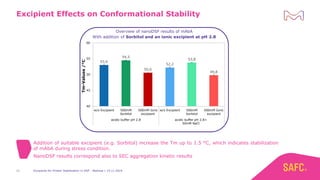
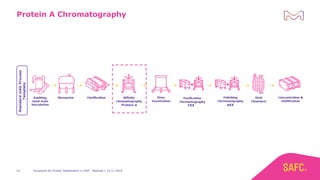
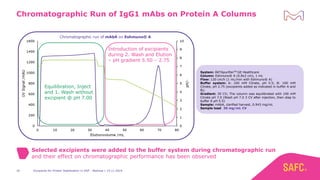
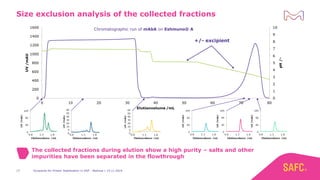
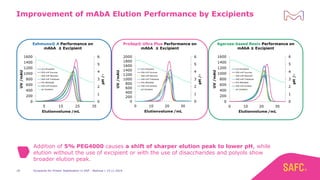
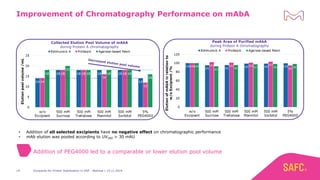
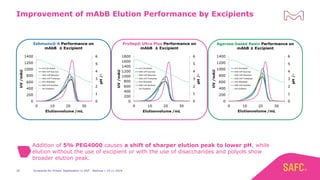
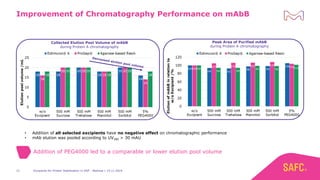
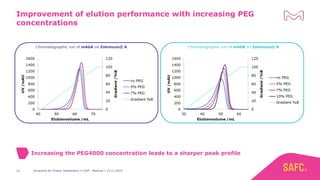
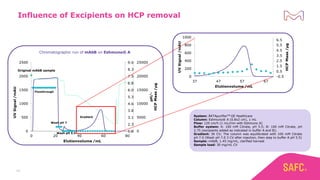
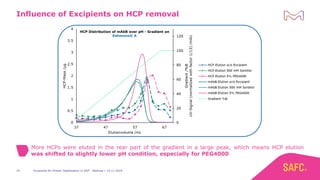
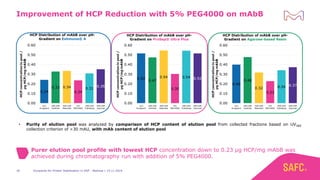
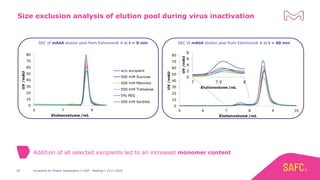
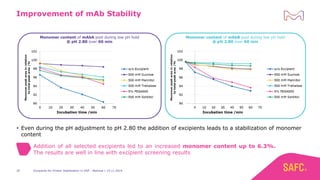
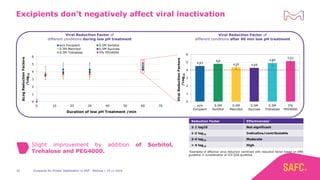
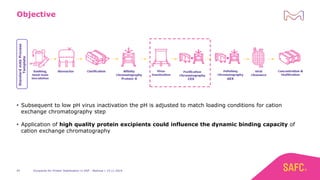
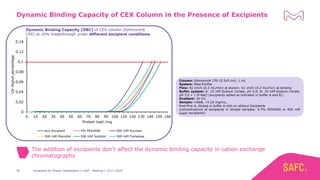
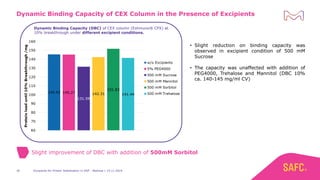
The webinar discusses the importance of excipients during downstream processing (DSP) of monoclonal antibodies (mAbs) to improve protein stability and solubility. Various excipients, including sugars, polyols, and surfactants, are tested for their effects on mAb aggregation and stability under acidic conditions, demonstrating significant stabilization effects. Additionally, the study highlights the successful use of selected excipients during protein A chromatography and virus inactivation processes, confirming no negative impact on viral inactivation rates.