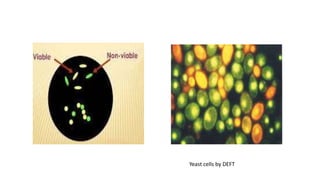
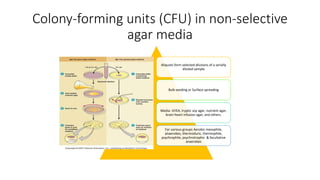
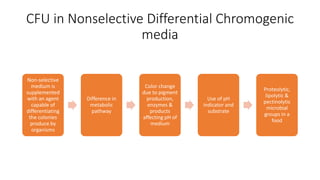
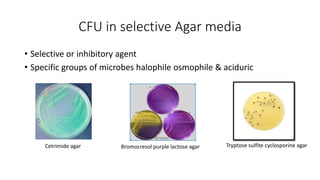
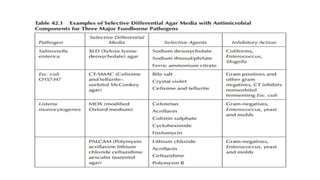
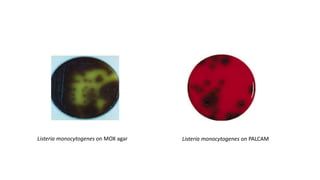
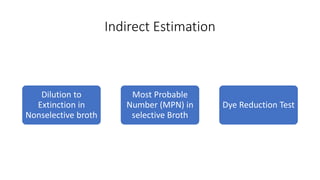
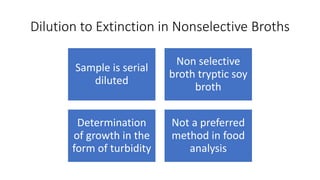
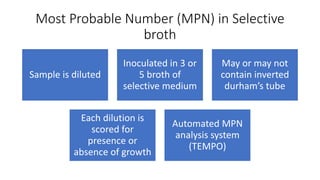
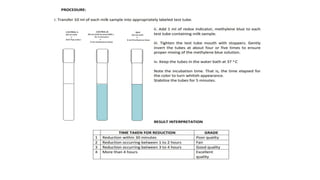
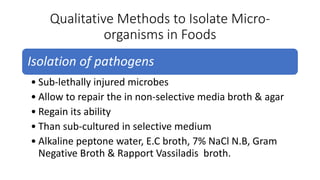
This document discusses conventional methods for sampling and analyzing food for microbial content. It covers both quantitative methods like colony forming units (CFU) on selective and differential media to enumerate microbes, and qualitative methods like isolation and toxin detection. Standard methods are outlined from regulatory publications. Considerations for sampling include sample size and plans to determine acceptance of food batches. Rapid automated methods are also discussed.