chemistary of nucleic acid-T.pdf
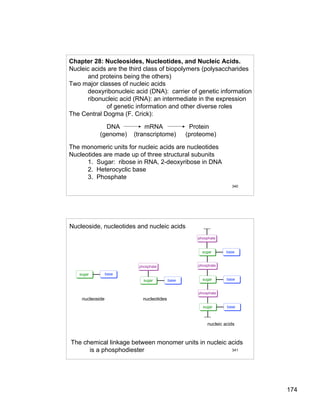
- 1. 174 340 Chapter 28: Nucleosides, Nucleotides, and Nucleic Acids. Nucleic acids are the third class of biopolymers (polysaccharides and proteins being the others) Two major classes of nucleic acids deoxyribonucleic acid (DNA): carrier of genetic information ribonucleic acid (RNA): an intermediate in the expression of genetic information and other diverse roles The Central Dogma (F. Crick): DNA mRNA Protein (genome) (transcriptome) (proteome) The monomeric units for nucleic acids are nucleotides Nucleotides are made up of three structural subunits 1. Sugar: ribose in RNA, 2-deoxyribose in DNA 2. Heterocyclic base 3. Phosphate 341 sugar base sugar base phosphate sugar base phosphate sugar base phosphate sugar base phosphate nucleoside nucleotides nucleic acids Nucleoside, nucleotides and nucleic acids The chemical linkage between monomer units in nucleic acids is a phosphodiester
- 2. 175 342 28.1: Pyrimidines and Purines. The heterocyclic base; there are five common bases for nucleic acids (Table 28.1, p. 1166). Note that G, T and U exist in the keto form (and not the enol form found in phenols) N N H N N N N purine pyrimidine N N H N N N N H N NH NH2 O NH2 N H N O NH2 N H NH O O N H NH O O H3C adenine (A) DNA/RNA guanine (G) DNA/RNA cytosine (C) DNA/RNA thymine (T) DNA uracil (U) RNA 1 2 3 4 5 6 7 8 9 1 2 3 4 5 6 28.2: Nucleosides. N-Glycosides of a purine or pyrimidine heterocyclic base and a carbohydrate. The C-N bond involves the anomeric carbon of the carbohydrate. The carbohydrates for nucleic acids are D-ribose and 2-deoxy-D-ribose 343 Nucleosides = carbohydrate + base (Table 28.2, p. 1168) ribonucleosides or 2’-deoxyribonucleosides O HO N HO N N N X NH2 O HO N HO N N NH X O NH2 RNA: X= OH, adenosine (A) DNA: X= H, 2'-deoxyadenosine (dA) RNA: X= OH, guanosine (G) DNA: X= H, 2'-deoxyguanosine (dG) O HO HO X N N O NH2 O HO HO N NH O O H3C RNA: X= OH, cytidine (C) DNA: X= H, 2'-deoxycytidine (dC) DNA: thymidine (T) O HO HO N NH O O RNA: R= H, uridine (U) 1 2 3 4 5 6 7 8 9 1' 2' 3' 4' 5' OH 1' 2' 3' 4' 5' 2 1 3 4 5 6 To differentiate the atoms of the carbohydrate from the base, the position number of the carbohydrate is followed by a ´ (prime). The stereochemistry of the glycosidic bond found in nucleic acids is β.
- 3. 176 344 28.3: Nucleotides. Phosphoric acid esters of nucleosides. Nucleotides = nucleoside + phosphate O HO X HO B ribonucleoside (X=OH) deoxyribonucleoside (X=H) ribonucleotide 5'-monophosphate (X=OH, NMP) deoxyribonucleotide 5'-monophosphate (X=H, dNMP) O HO X O B P HO O O O HO X O B P O O O P HO O O O HO X O B P O O O P O O O P HO O O ribonucleotide 5'-diphosphate (X=OH, NDP) deoxyribonucleotide 5'-diphosphate (X=H, dNDP) ribonucleotide 5'-triphosphate (NTP) deoxyribonucleotide 5'-triphosphate (X=H, dNTP) O O OH O B P O O ribonucleotide 3',5'-cyclic phosphosphate (cNMP) Kinase: enzymes that catalyze the phosphoryl transfer reaction from ATP to an acceptor substrate. M2+ dependent O HO HO OH OH OH O N N N N NH2 HO OH O P O O O P O O O P O O O O HO HO OH OH OPO3 2- ATP O N N N N NH2 HO OH O P O O O P O O O ADP Glucose-6-phosphate Glucose 345 O N N N N NH2 HO OH O P O O O P O O O P O O O ATP O N N N N NH2 HO OH O P O O O AMP O O OH O P O O N N N N NH2 -P2O7 adenylyl cyclase H2O cAMP O N N N NH O HO OH O P O O O P O O O P O O O GTP O N N N NH O HO OH O P O O O GMP O O OH O P O O N N N NH O -P2O7 guanylate cyclase H2O cGMP NH2 NH2 NH2 1971 Nobel Prize in Medicine or Physiology: Earl Sutherland 28.4: Bioenergetics. (Please read) 28.5: ATP and Bioenergetics. (Please read) + HPO4 2- O N N N N NH2 HO OH O P O O O P O O O P O O O ATP H2O O N N N N NH2 HO OH O P O O O P O O O ADP + ~31 KJ/mol (7.4 Kcal/mol)
- 4. 177 346 28.6: Phosphodiesters, Oligonucleotides, and Polynucleotides. The chemical linkage between nucleotide units in nucleic acids is a phosphodiester, which connects the 5’-hydroxyl group of one nucleotide to the 3’-hydroxyl group of the next nucleotide. By convention, nucleic acid sequences are written from left to right, from the 5’-end to the 3’-end. Nucleic acids are negatively charged 28.7: Nucleic Acids. 1944: Avery, MacLeod & McCarty - Strong evidence that DNA is genetic material 1950: Chargaff - careful analysis of DNA from a wide variety of organisms. Content of A,T, C & G varied widely according to the organism, however: A=T and C=G (Chargaff’ Rule) 1953: Watson & Crick - structure of DNA (1962 Nobel Prize with M. Wilkens, 1962) RNA, X= OH DNA, X=H O O O X Base P O O O O O X Base P O O O 3' 3' 5' 5' 3' 5' 347 28.8: Secondary Structure of DNA: The Double Helix. Initial “like-with-like”, parallel helix: Does not fit with with Chargaff’s Rule: A = T G = C Wrong tautomers !! Watson, J. D. The Double Helix, 1968 N N N N N H dR N N N N N H dR H N N dR O H O N N dR O H O purine - purine pyrimidine - pyrimidine N N dR N H O N N dR N H O H N N N N O N H dR N N N N O H dR H H H N H H H
- 5. 178 348 Two polynucleotide strands, running in opposite directions (anti-parallel) and coiled around each other in a double helix. The strands are held together by complementary hydrogen- bonding between specific pairs of bases. O OR RO N N N O H H O OR RO N N N N O N H H H Hydrogen Bond Donor Hydrogen Bond Donor Hydrogen Bond Acceptor Hydrogen Bond Acceptor 3' 5' Hydrogen Bond Acceptor Hydrogen Bond Donor 5' O2 N4 O6 3' Antiparallel C-G Pair N3 N1 N2 O OR RO N N O O O OR RO N N N N N H H H 5' 3' 3' 5' Hydrogen Bond Donor N6 Hydrogen Bond Acceptor Hydrogen Bond Acceptor O4 Hydrogen Bond Donor Antiparallel T-A Pair N1 N3 349 DNA double helix one helical turn 34 Å major groove 12 Å minor groove 6 Å backbone: deoxyribose and phosphodiester linkage bases
- 6. 179 350 28.9: Tertiary Structure of DNA: Supercoils. Each cell contains about two meters of DNA. DNA is “packaged” by coiling around a core of proteins known as histones. The DNA-histone assembly is called a nucleosome. Histones are rich is lysine and arginine residues. Pdb code 1kx5 351 28.10: Replication of DNA. The Central Dogma (F. Crick): DNA replication DNA transcription mRNA translation Protein (genome) (transcriptome) (proteome) Expression and transfer of genetic information: Replication: process by which DNA is copied with very high fidelity. Transcription: process by which the DNA genetic code is read and transferred to messenger RNA (mRNA). This is an intermediate step in protein expression Translation: The process by which the genetic code is converted to a protein, the end product of gene expression. The DNA sequence codes for the mRNA sequence, which codes for the protein sequence “It has not escaped our attention that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material.” Watson & Crick
- 7. 180 352 DNA is replicated by the coordinated efforts of a number of proteins and enzymes. For replication, DNA must be unknotted, uncoiled and the double helix unwound. Topoisomerase: Enzyme that unknots and uncoils DNA Helicase: Protein that unwinds the DNA double helix. DNA polymerase: Enzyme that replicates DNA using each strand as a template for the newly synthesized strand. DNA ligase: enzyme that catalyzes the formation of the phosphodiester bond between pieces of DNA. DNA replication is semi-conservative: Each new strand of DNA contains one parental (old, template) strand and one daughter (newly synthesized) strand 353 Unwinding of DNA by helicases expose the DNA bases (replication fork) so that replication can take place. Helicase hydrolyzes ATP in order to break the hydrogen bonds between DNA strands DNA replication
- 8. 181 354 DNA Polymerase: the new strand is replicated from the 5’ → 3’ (start from the 3’-end of the template) DNA polymerases are Mg2+ ion dependent The deoxynucleotide 5’-triphosphate (dNTP) is the reagent for nucleotide incorporation G O O O P O -O A O O O T O OH O P O O C O OH O O- P O- O O P O- O O- Mg2+ 5' 3' 5' template strand (old) (new) 3’-hydroxyl group of the growing DNA strand acts as a nucleophile and attacks the α-phosphorus atom of the dNTP. dNTP 355 Replication of the leading strand occurs continuously in the 5’ → 3’ direction of the new strand. Replication of the lagging strand occurs discontinuously. Short DNA fragments are initially synthesized and then ligated together. DNA ligase catalyzes the formation of the phosphodiester bond between pieces of DNA. animations of DNA processing: http://www.wehi.edu.au/education/wehi-tv/dna/
- 9. 182 356 DNA replication occurs with very high fidelity: Most DNA polymerases have high intrinsic fidelity Many DNA polymerases have “proof-reading” (exonuclease) activity Mismatch repair proteins seek out and repair base-pair mismatches due to unfaithful replication 28.11 Ribonucleic Acid RNA contains ribose rather than 2-deoxyribose and uracil rather than thymine. RNA usually exist as a single strand. There are three major kinds of RNA messenger RNA (mRNA): ribosomal RNA (rRNA) transfer RNA (tRNA) DNA is found in the cell nucleus and mitochondria; RNA is more disperse in the cell. 357 Transcription: only one of the DNA strands is copied (coding or antisense strand). An RNA polymerase replicates the DNA sequence into a complementary sequence of mRNA (template or sense strand). mRNAs are transported from the nucleus to the cytoplasm, where they acts as the template for protein biosynthesis (translation). A three base segment of mRNA (codon) codes for an amino acid.The reading frame of the codons is defined by the start and stop codons.
- 10. 183 358 5'-cap 5'-UTR start stop 3'-UTR Coding sequence The mRNA is positioned in the ribosome through complementary pairing of the 5’-untranslated region of mRNA with a rRNA. Transfer RNA (tRNA): t-RNAs carries an amino acid on the 3’-terminal hydroxyl (A) (aminoacyl t-RNA) and the ribosome catalyzes amide bond formation. Ribosome: large assembly of proteins and rRNAs that catalyzes protein and peptide biosynthesis using specific, complementary, anti-parallel pairing interactions between mRNA and the anti-codon loop of specific tRNA’s. 359 Although single-stranded, there are complementary sequences within tRNA that give it a defined conformation. The three base codon sequence of mRNA are complementary to the “anti-codon” loops of the appropriate tRNA. The base- pairing between the mRNA and the tRNA positions the tRNAs for amino acid transfer to the growing peptide chain. aminoacyl t-RNA TψC loop D loop variable loop
- 11. 184 360 28.12: Protein Biosynthesis. Ribosomal protein synthesis P site U G U A A U C U C G U U 3' 5' A site A C U O O H N SCH3 OHC U G U A A U C U C G U U 3' 5' A C U O O H N SCH3 OHC U A A O O H2N OH U G U A A U C U C G U U 3' 5' U A A O O HN OH O N H CH3S A C U OH OHC U G U A A U C U C G U U 3' 5' U A A O O HN OH O N H CH3S A C U OH OHC U G U A A U C U C G U U 3' 5' U A A O O HN OH O N H CH3S A C U OH OHC E site E site U G U A A U C U C G U U 3' 5' U A A O O HN OH O N H CH3S OHC G A C O O CH3 H2N U G U A A U C U C G U U 3' 5' O HN OH O HN G A C O O CH3 HN SCH3 U A A OH OJHC U G U A A U C U C G U U 3' 5' O HN OH O HN G A C O O CH3 HN SCH3 U A A OH OHC U G U A A U C U C G U U 3' 5' O HN OH O HN G A C O O CH3 HN SCH3 U A A OH OHC 361 28.13: AIDS. (please read) 28.14: DNA Sequencing. Maxam-Gilbert: relys on reagents that react with a specific DNA base that can subsequent give rise to a sequence specific cleavage of DNA Sanger: Enzymatic replication of the DNA fragment to be sequenced with a DNA polymerase, Mg+2, and dideoxynucleotides triphosphate (ddNTP) that truncates DNA replication Restriction endonucleases: Bacterial enzymes that cleave DNA at specific sequences 5’-d(G-A-A-T-T-C)-3’ 3’-d(C-T-T-A-A-G)-5’ 5’-d(G-G-A-T-C-C)-3’ 3’-d(C-C-T-A-G-G)-5’ EcoR I BAM HI 5' 3' 3' 5' 5' 3' 3' 5' OH 2-O3PO 2-O3PO OH restriction enzyme
- 12. 185 362 Sanger Sequencing: key reagent: dideoxynucleotides triphosphates (ddNTP) N N N N NH2 O O P O O- O P O O- O P O -O O- NH N N N O O O P O O- O P O O- O P O -O O- ddATP NH2 ddGTP O O P O O- O P O O- O P O -O O- O O P O O- O P O O- O P O -O O- ddTTP ddCTP N NH O O N N NH2 O When a ddNTP is incorporated elongation of the primer is terminated The ddNTP is specifically incorporated opposite its complementary nucleotide base DNA polymerase 3' 5'-32P 3' 5' primer Template unknown sequence Mg2+, dNTP's + one ddATP O N3 HO N NH O O O HO N N N NH O S O OH N N H2N O O HO N N NH2 O ddI AZT (-)-3TC Anti-Viral Nucleosides d4C 363 Sanger Sequencing Larger fragments Smaller fragments G T A A C G T A A T C A C A G ddA ddG ddC ddT C A T T G C A T T A G T G T C 5' 32 P 3' 5' 3' 32 P-5' 3' primer template
- 13. 186 364 28.15: The Human Genome Project. (please read) 28.16: DNA Profiling and Polymerase Chain Reaction (PCR). method for amplifying DNA using a DNA polymerase, dNTPs and cycling the temperature. Heat stable DNA Polymerases (from archaea): Taq: thermophilic bacteria (hot springs)- no proof reading Pfu: geothermic vent bacteria- proof reading Mg 2+ two Primer DNA strands (synthetic, large excess) one sense primer and one antisense primer one Template DNA strand (double strand) dNTP’s 1 x 2 = 2 x 2 = 4 x 2 = 8 x 2 = 16 x 2 = 32 x 2 = 64 x 2 = 128 x 2 = 256 x 2 = 512 x 2 = 1,024 x 2 = 2,048 x 2 = 4,096 x 2 = 8,192 x 2 = 16,384 x 2 = 32,768 x 2 = 65,536 x 2 = 131,072 x 2 = 262,144 x 2 = 524,288 x 2 = 1,048,576 In principle, over one million copies per original, can be obtained after just twenty cycles KARY B. MULLIS, 1993 Nobel Prize in Chemistry for his invention of the polymerase chain reaction (PCR) method. 365 Polymerase Chain Reaction For a PCR animation go to: http://www.blc.arizona.edu/INTERACTIVE/recombinant3.dna/pcr.html http://users.ugent.be/~avierstr/principles/pcrani.html 5' 3' 3' 5' 95 °C denaturation 5' 3' 3' 5' anneal (+) and (-) primers 55 - 68 °C 5' 3' 3' 5' 3' 3' 72 °C Taq, Mg 2+, dNTPs extension 5' 3' 3' 5' 5' 3' 3' 5' 95 °C denaturation 2nd cycle 5' 3' 3' 5' 5' 3' 3' 5' amplification of DNA 2 copies of DNA repeat temperature cycles