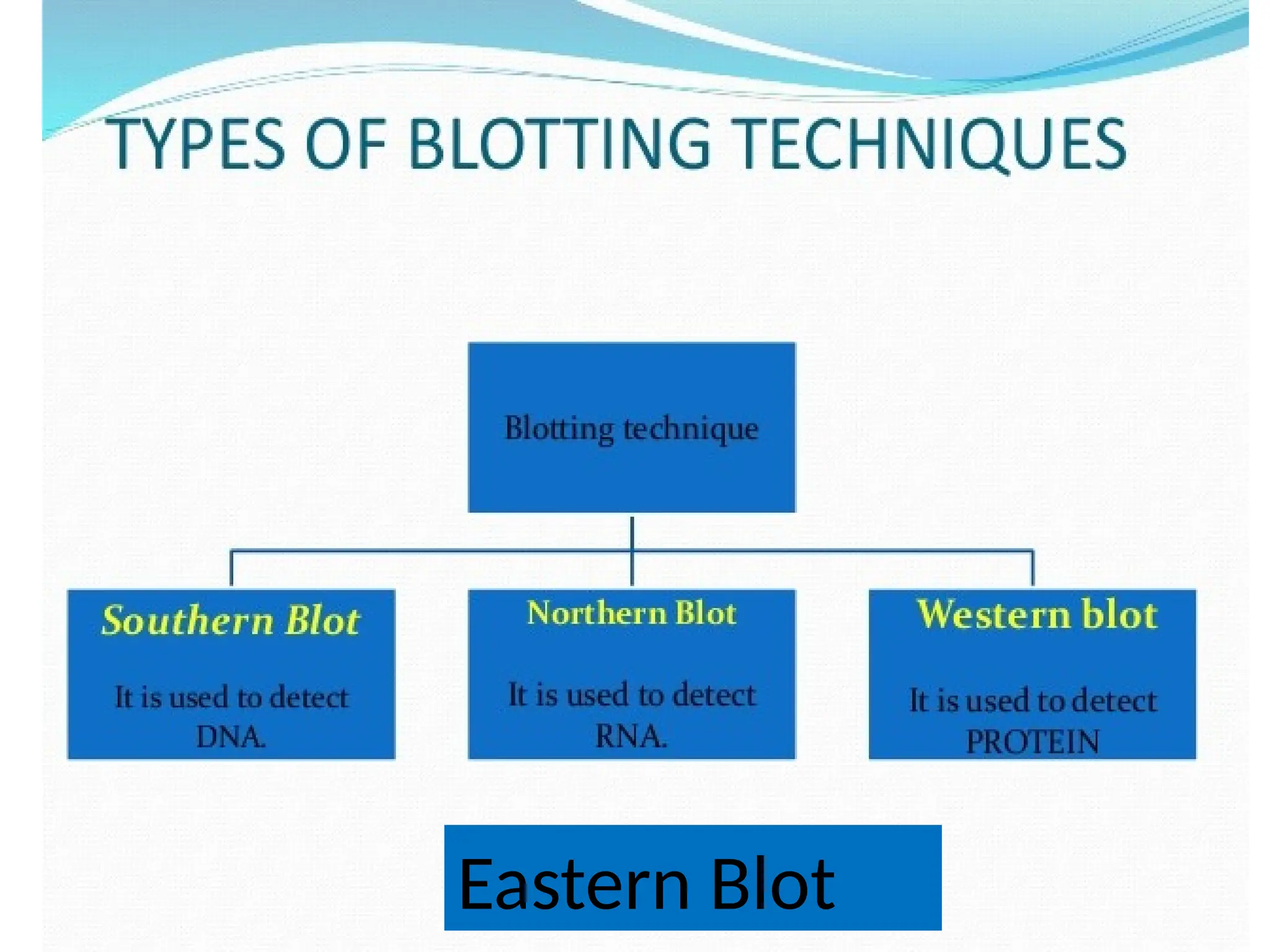
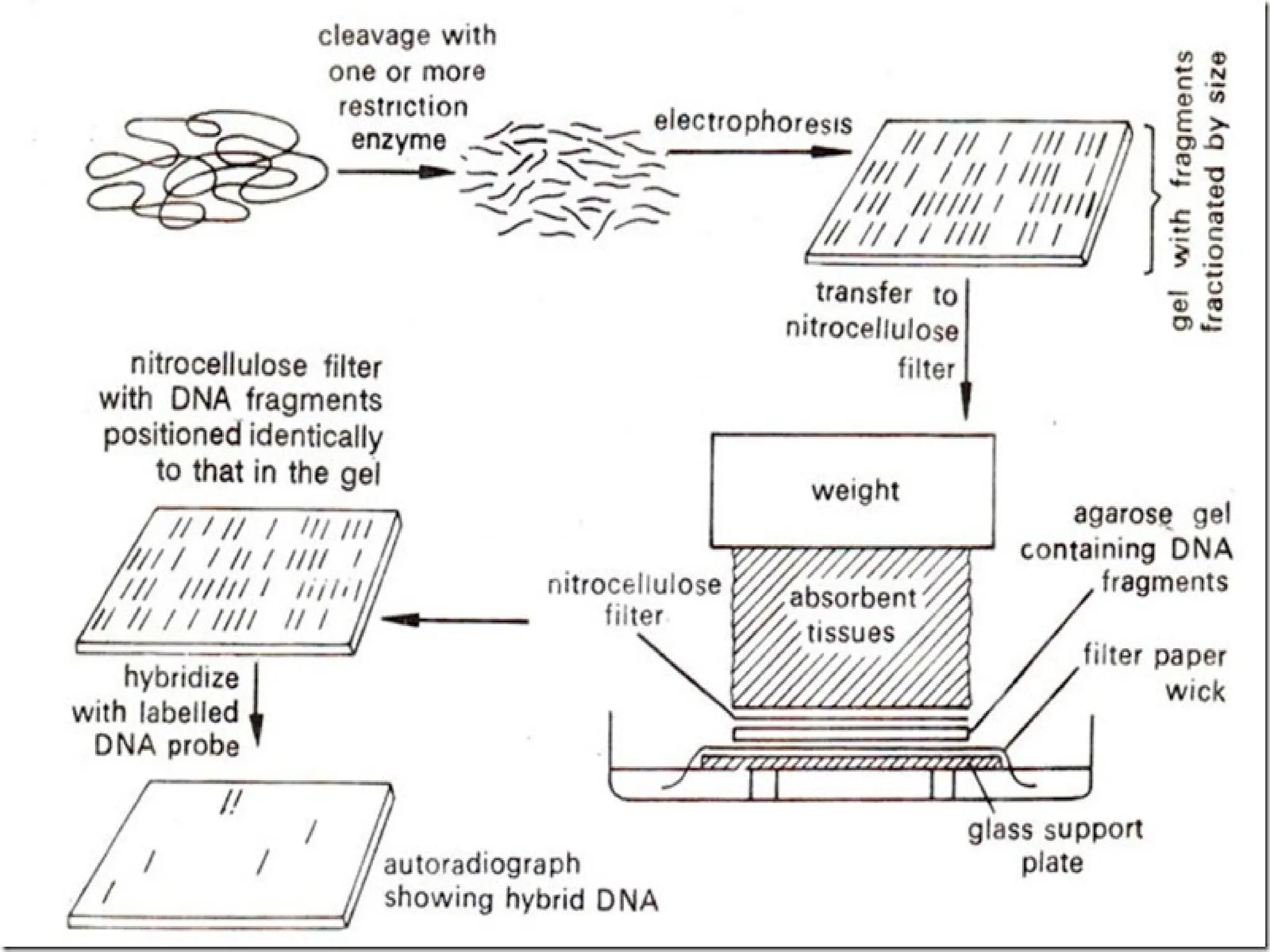
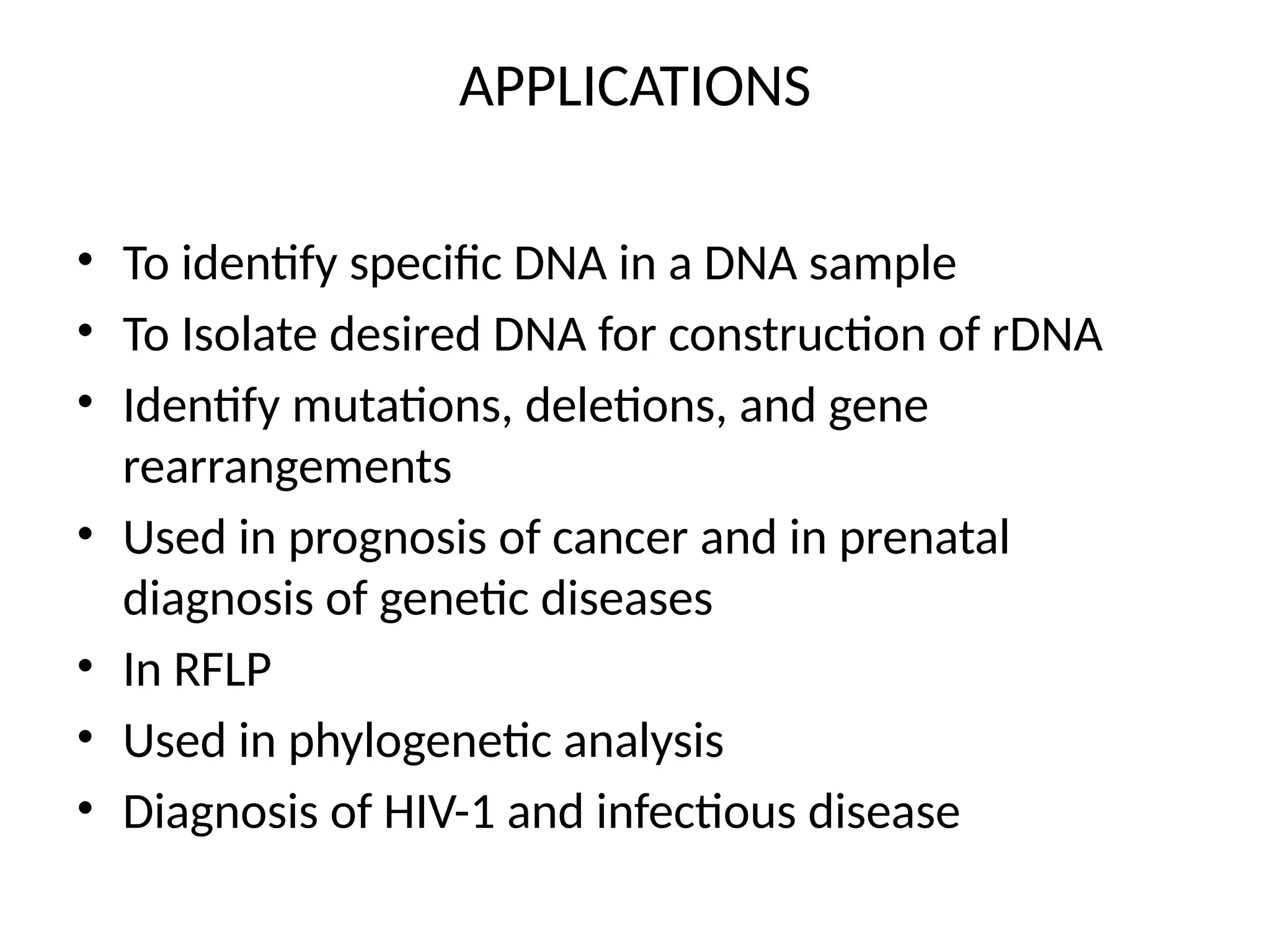
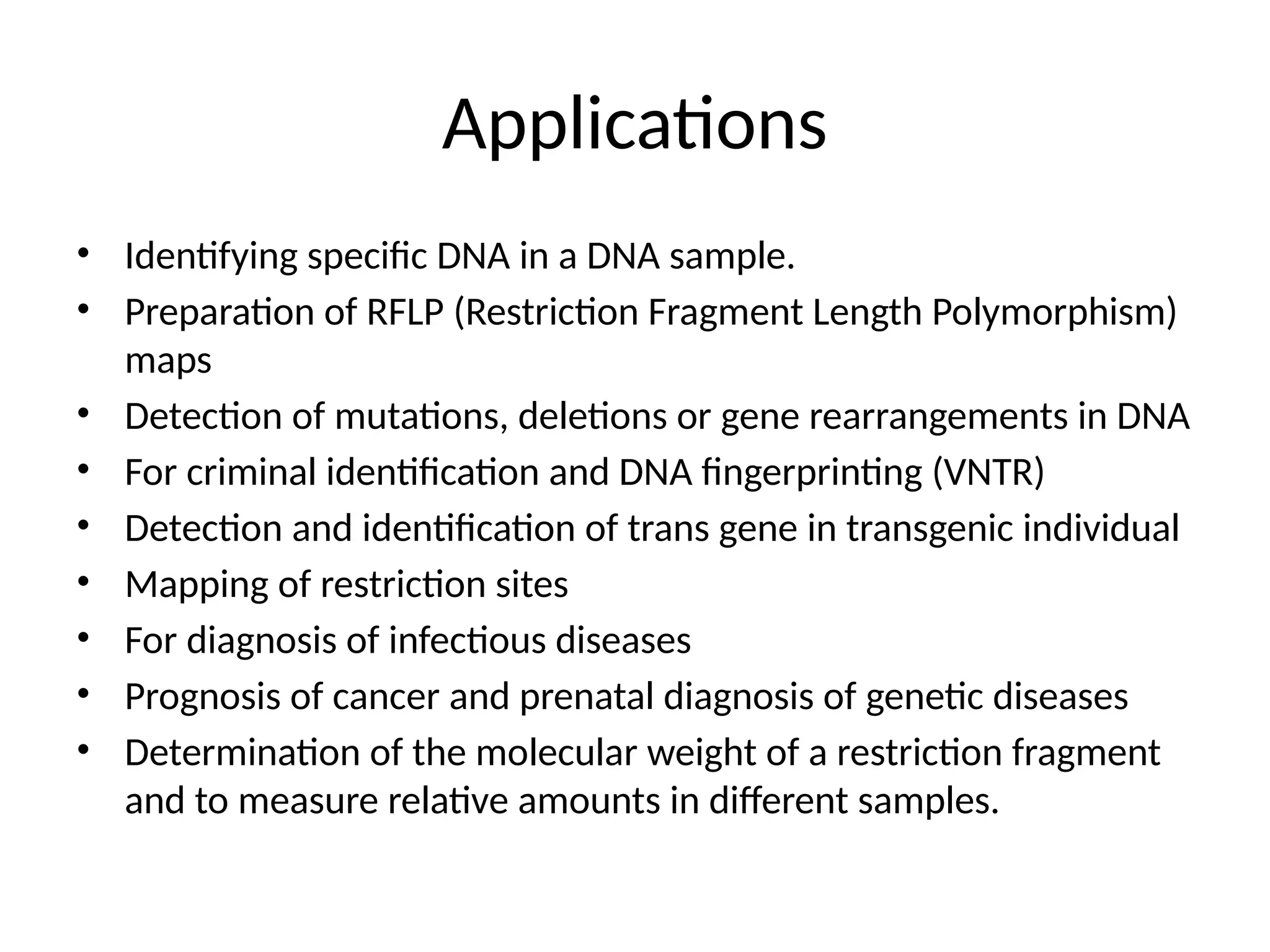
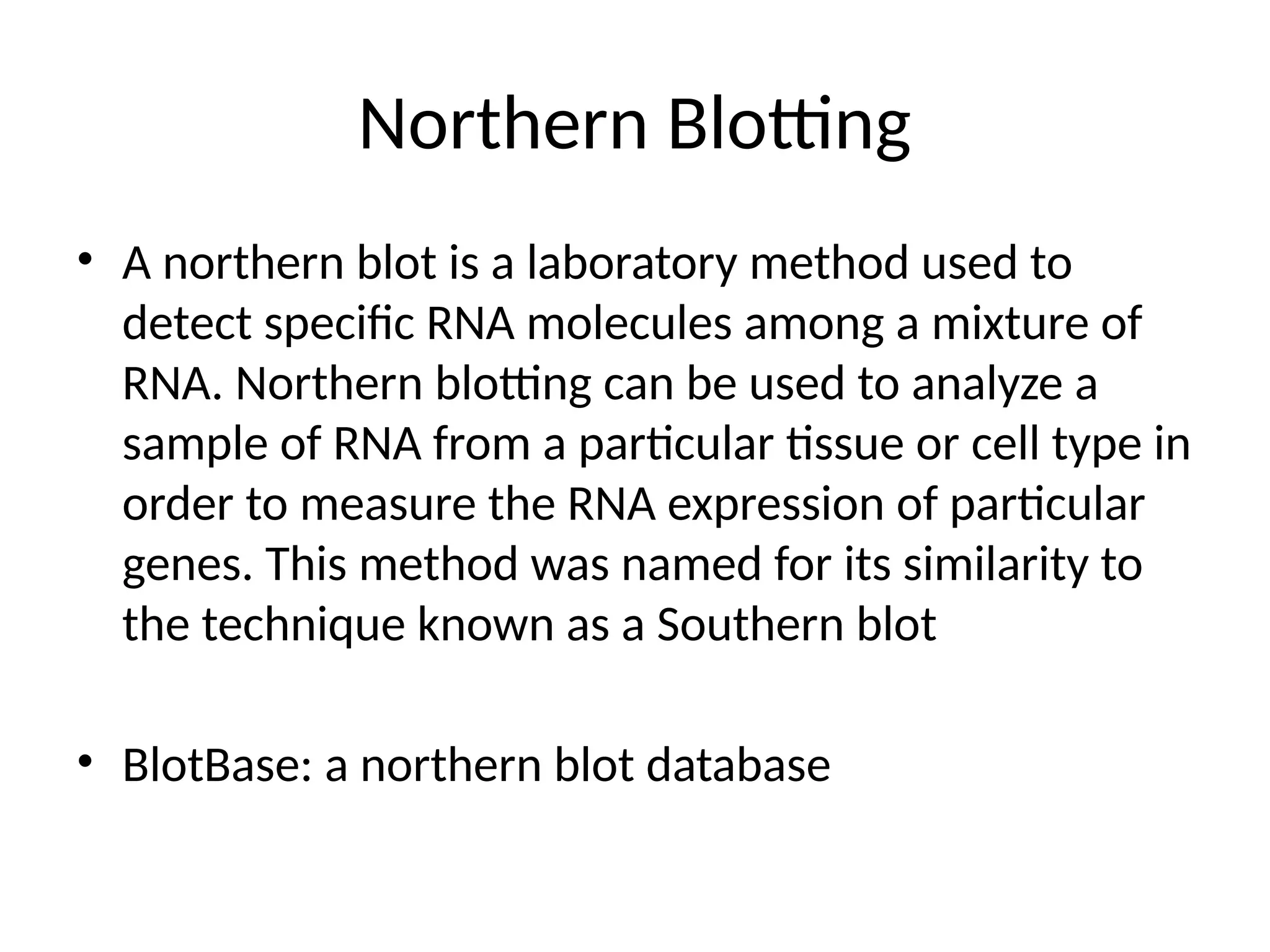
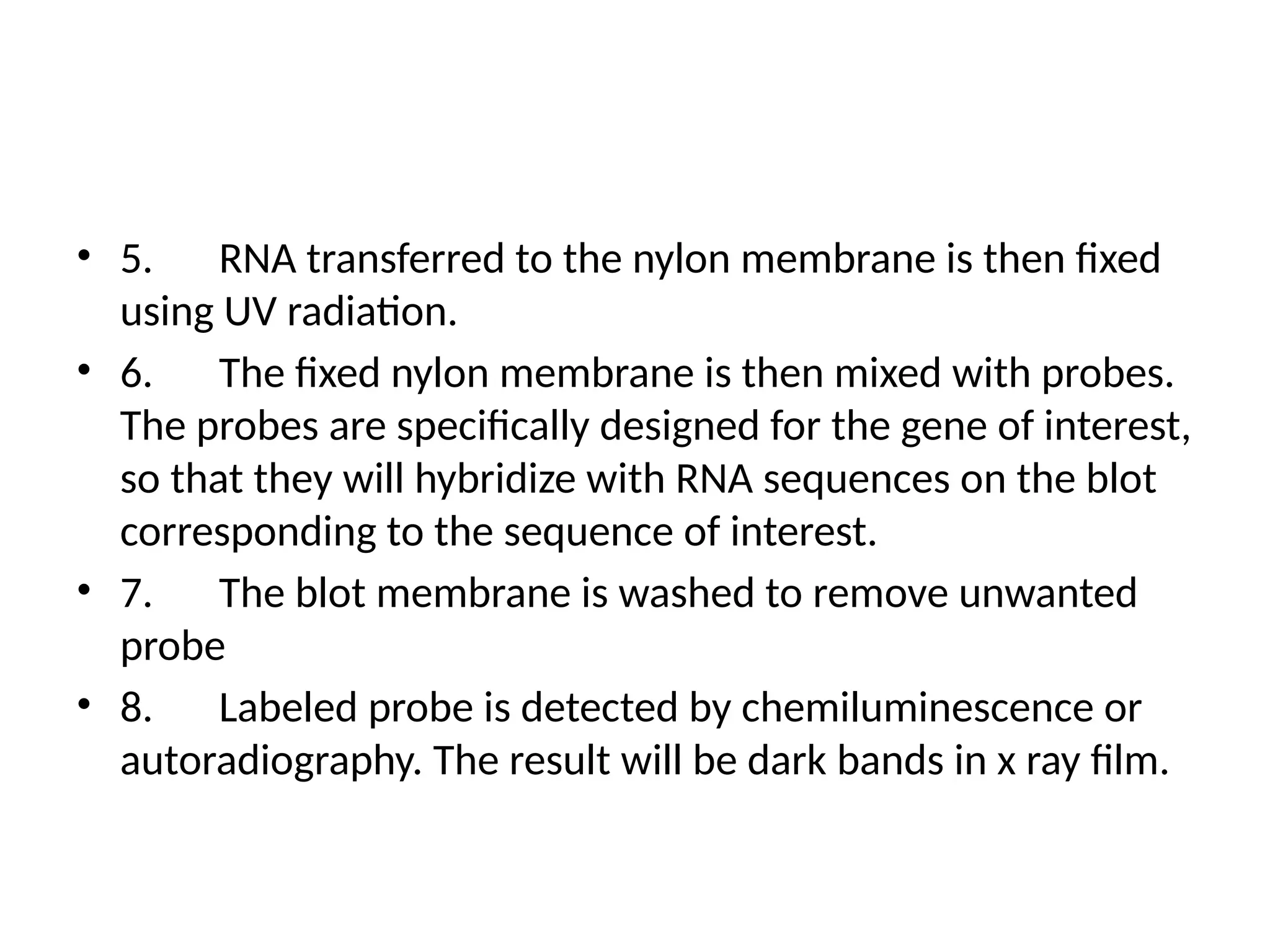
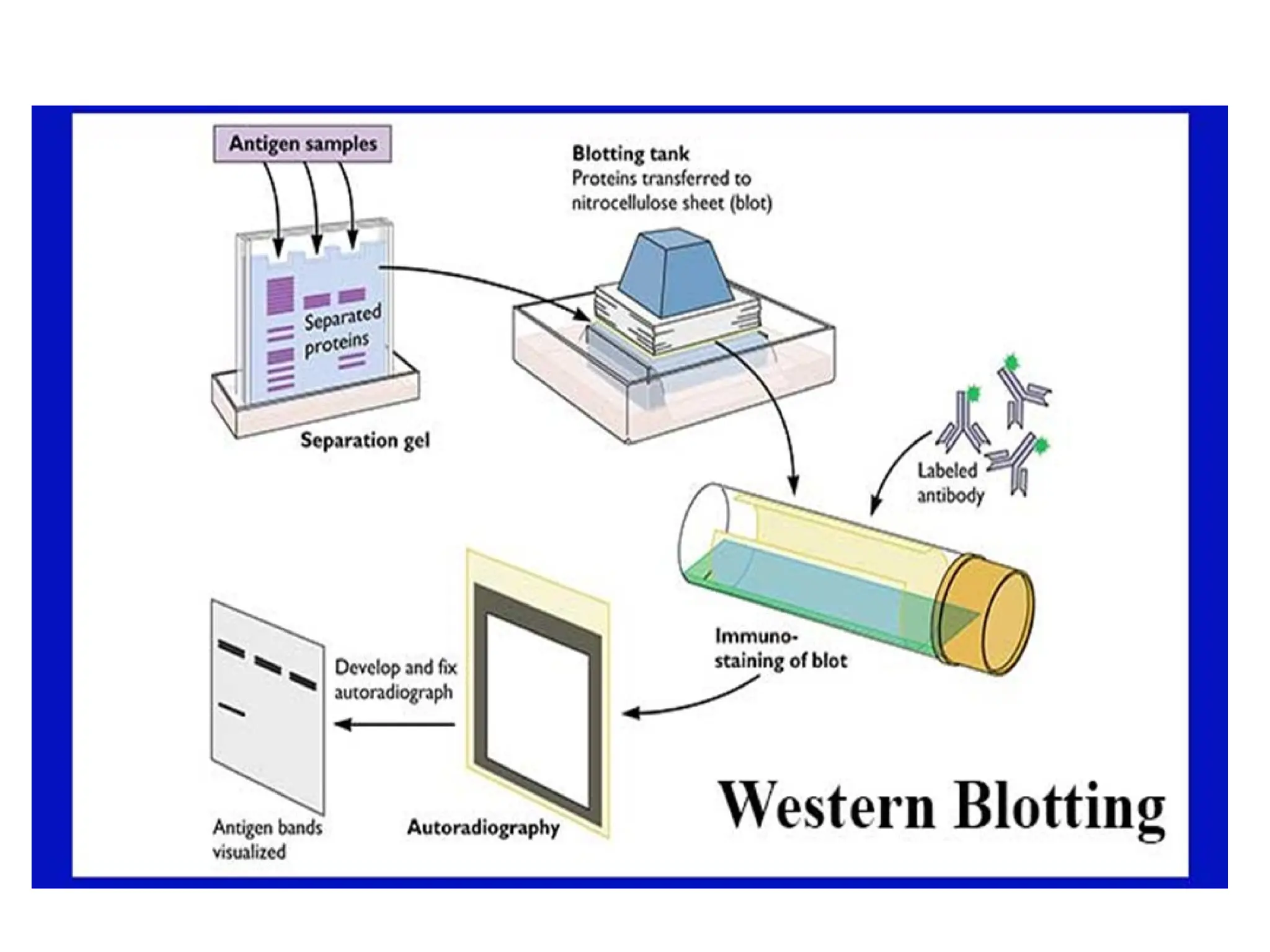
Blotting techniques, including Southern, Northern, and Western blotting, are crucial methods for transferring and detecting DNA, RNA, and proteins, respectively. These techniques involve separation by electrophoresis, transfer to a membrane, and detection via specific probes, with a variety of applications such as genetic analysis, disease diagnosis, and forensic identification. Each method has distinct processes and uses probes for specific target molecules to visualize results.