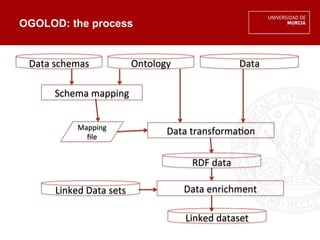
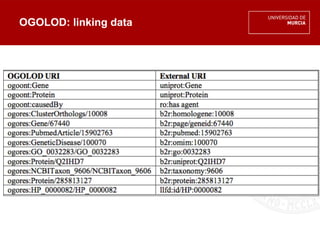
The document discusses the integration of multiple orthology databases to enhance research capabilities, particularly in linking genes to diseases and functional activities. It highlights existing resources and tools, such as ogolod and orthoxml, along with their implications for data sharing and ontology mapping. The author emphasizes the need for standardization in orthology resources and addresses challenges faced in the field during the Biohackathon 2014.