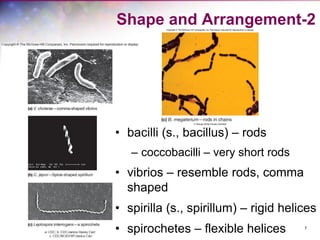
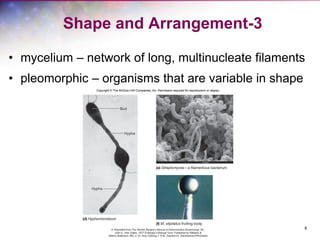
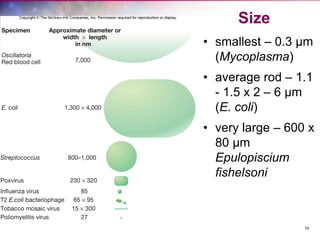
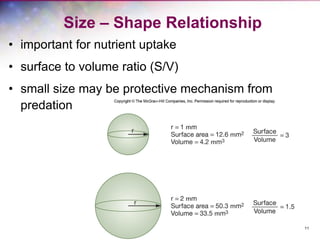
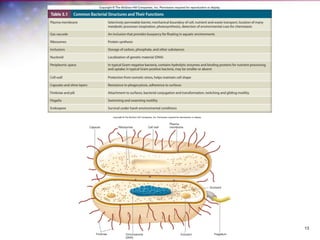
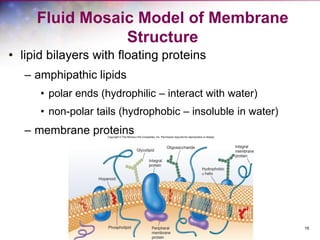
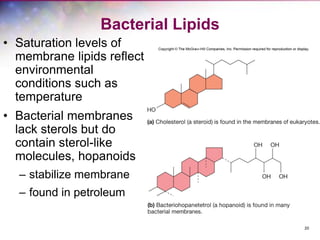
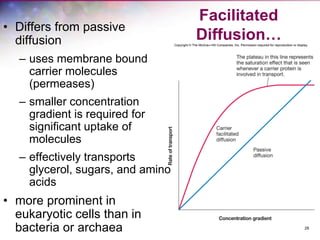
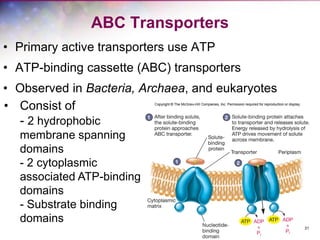
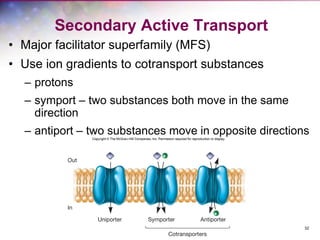
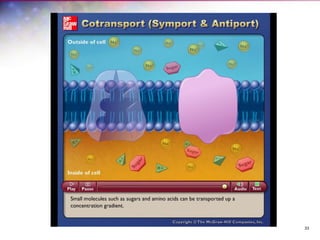
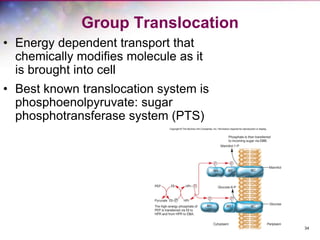
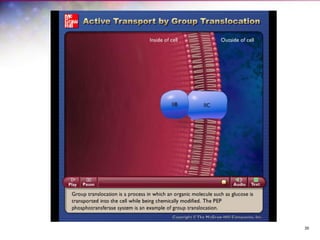
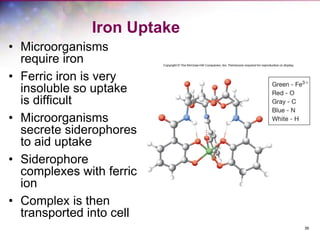
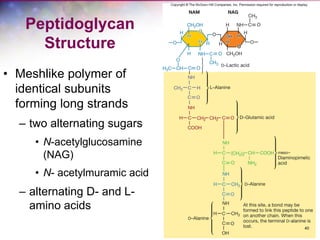
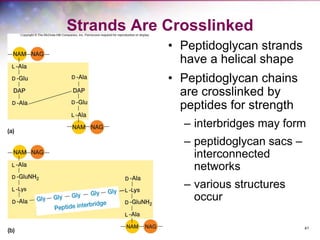
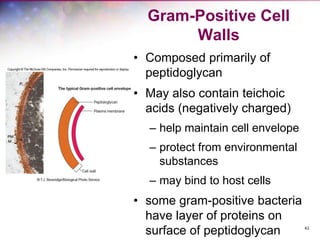
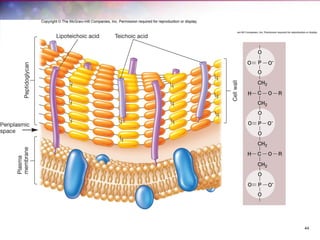
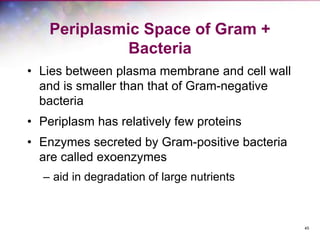
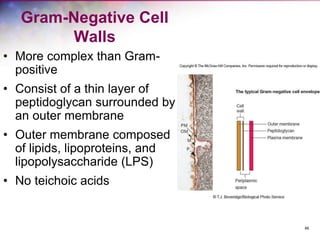
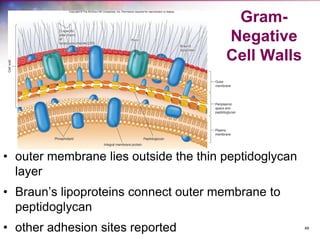
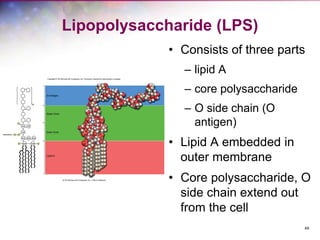
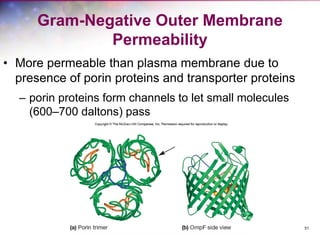
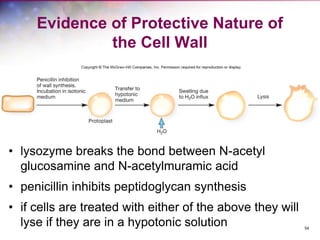
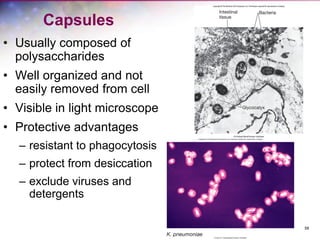
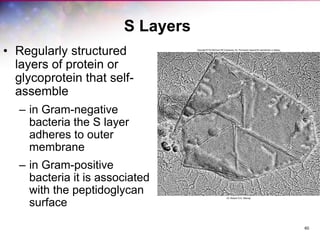
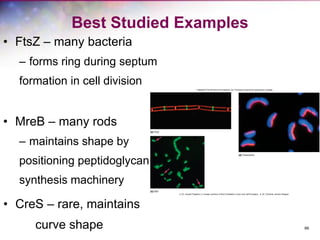
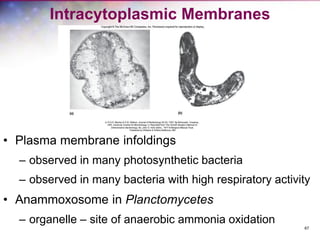
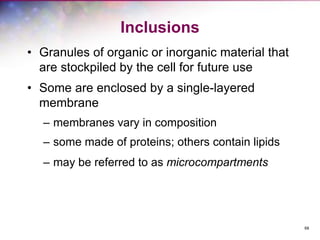
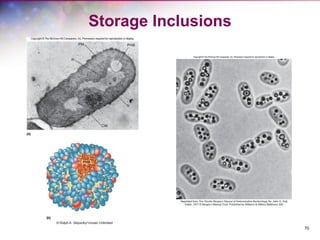
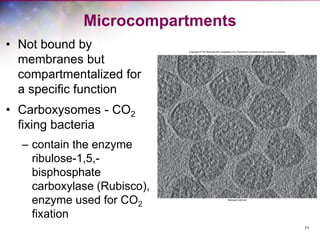
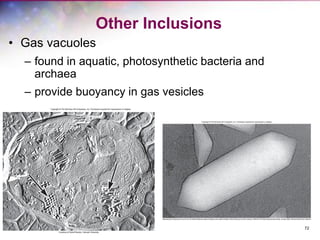
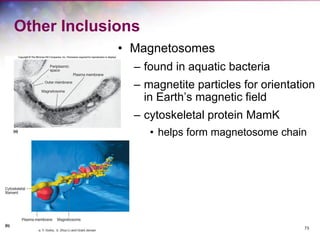
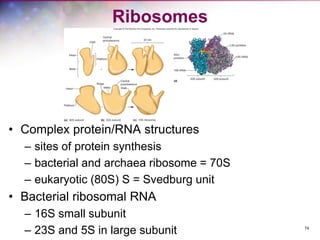
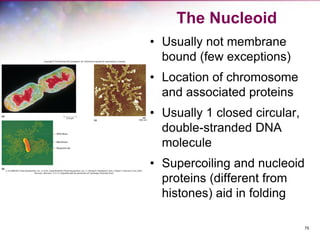
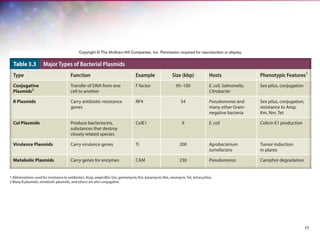
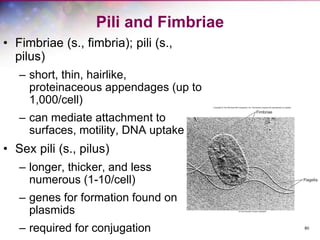
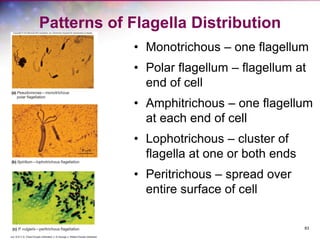
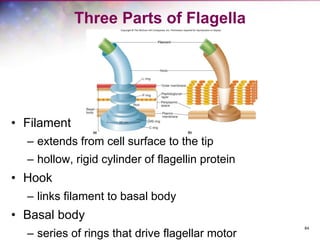
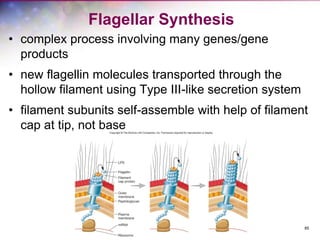
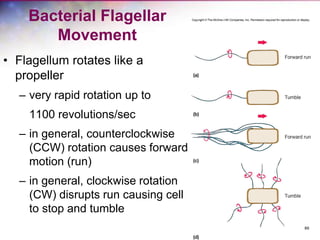
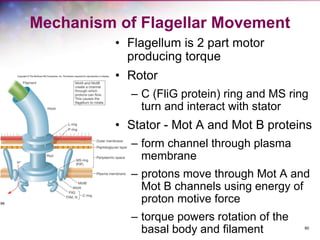
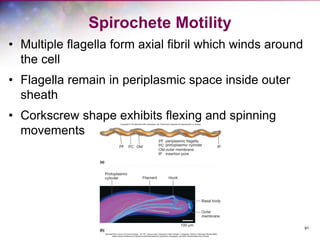
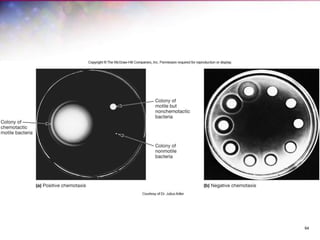
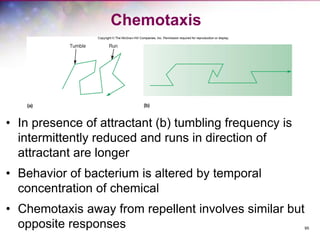
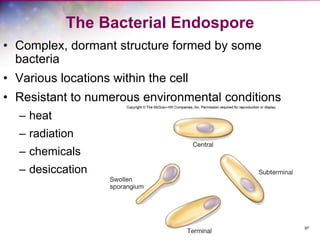
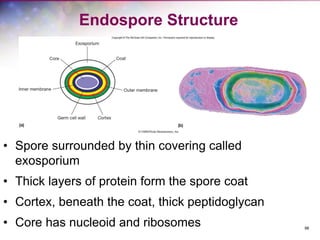
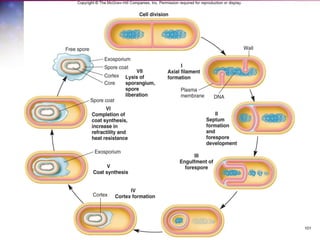
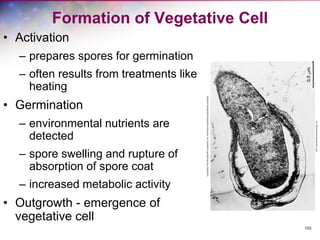
The document discusses the structure and function of bacterial cells, covering topics such as prokaryotic cell characteristics, cell size, shape, organization, and key components like the cell wall, plasma membrane, and cytoplasm. It emphasizes differences between gram-positive and gram-negative bacteria, including their cell wall structures and the gram-staining reaction. Additionally, it outlines mechanisms for nutrient uptake and the significance of external structures like pili and flagella for bacterial movement and attachment.