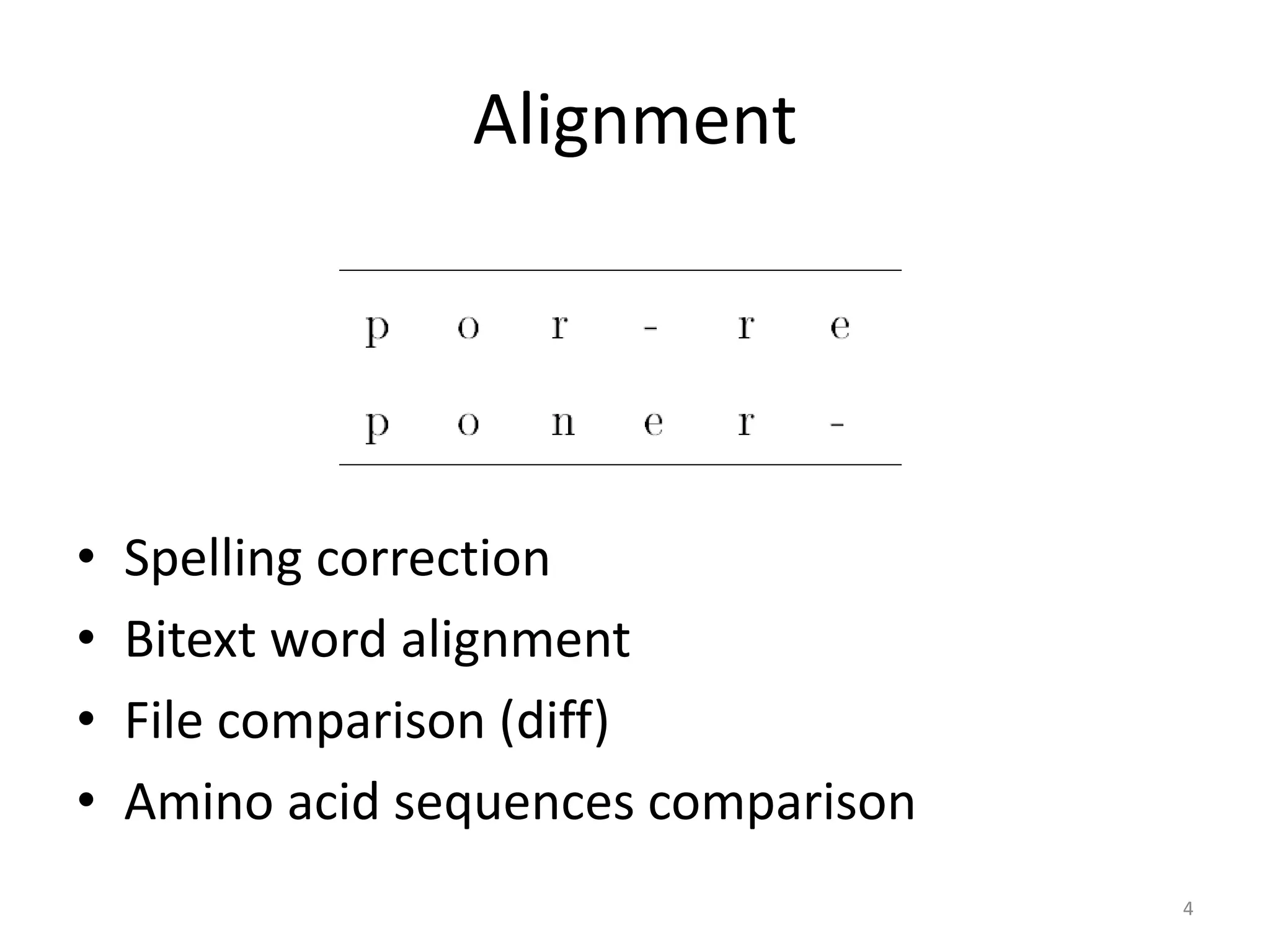
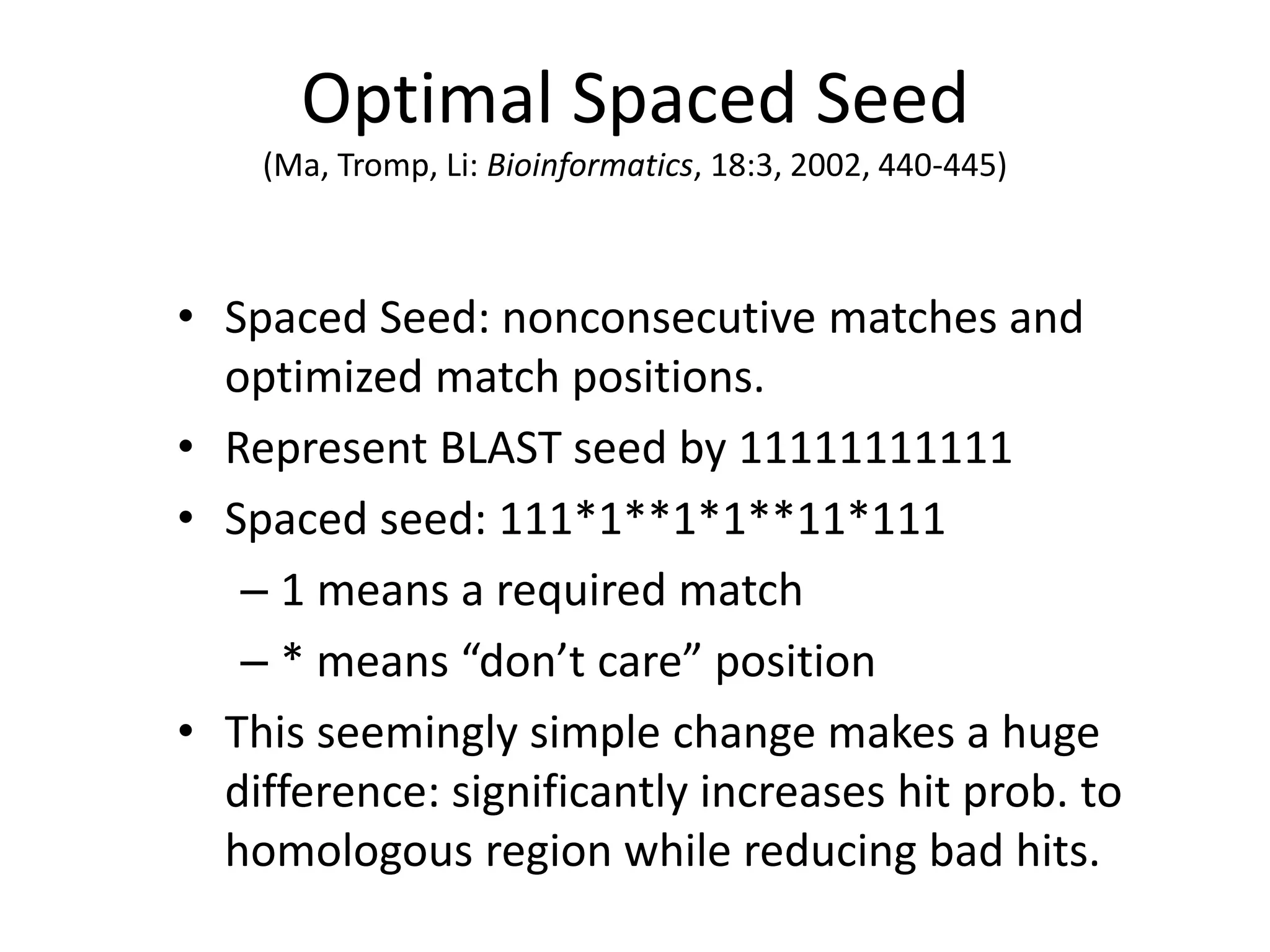
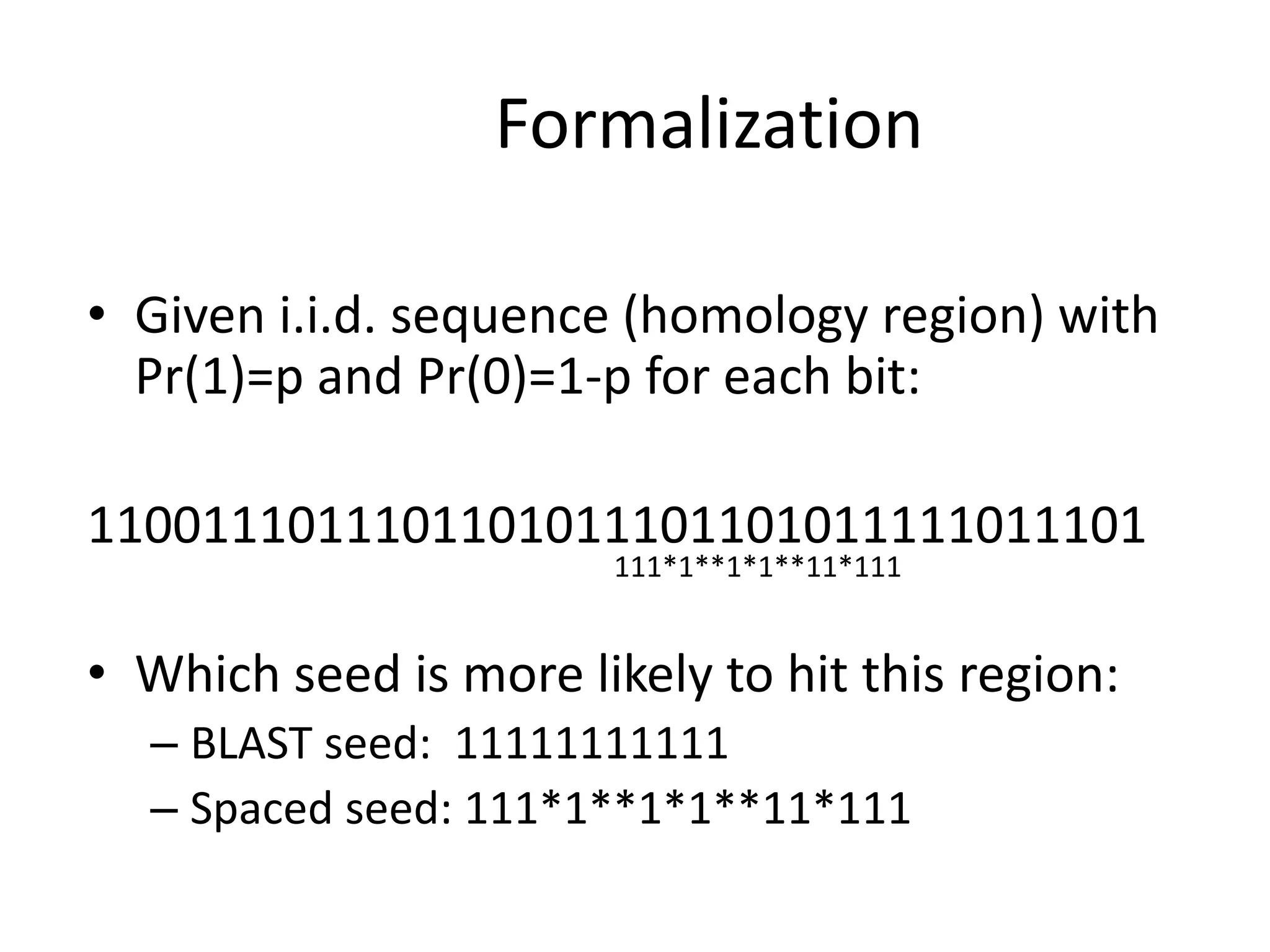
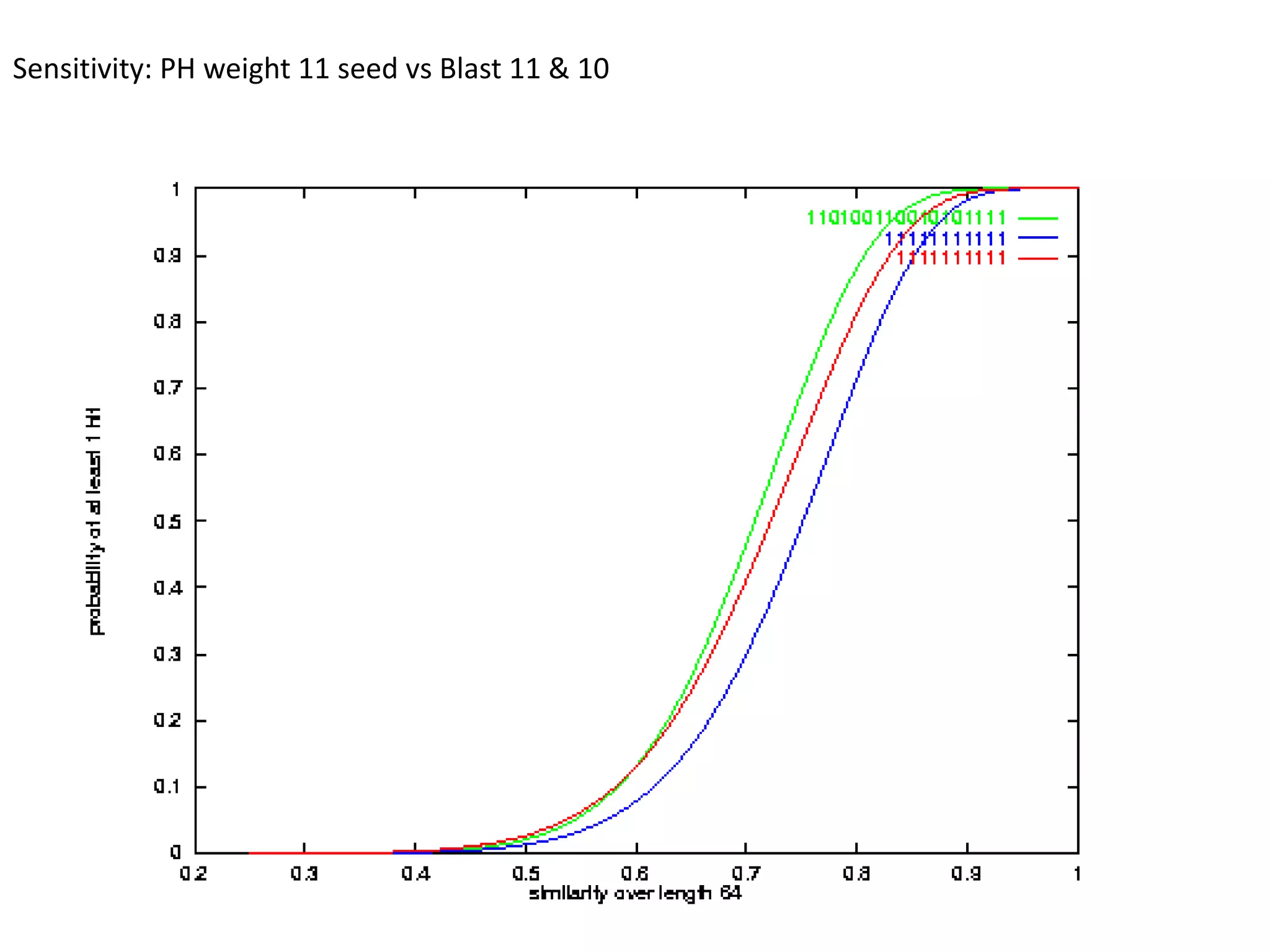
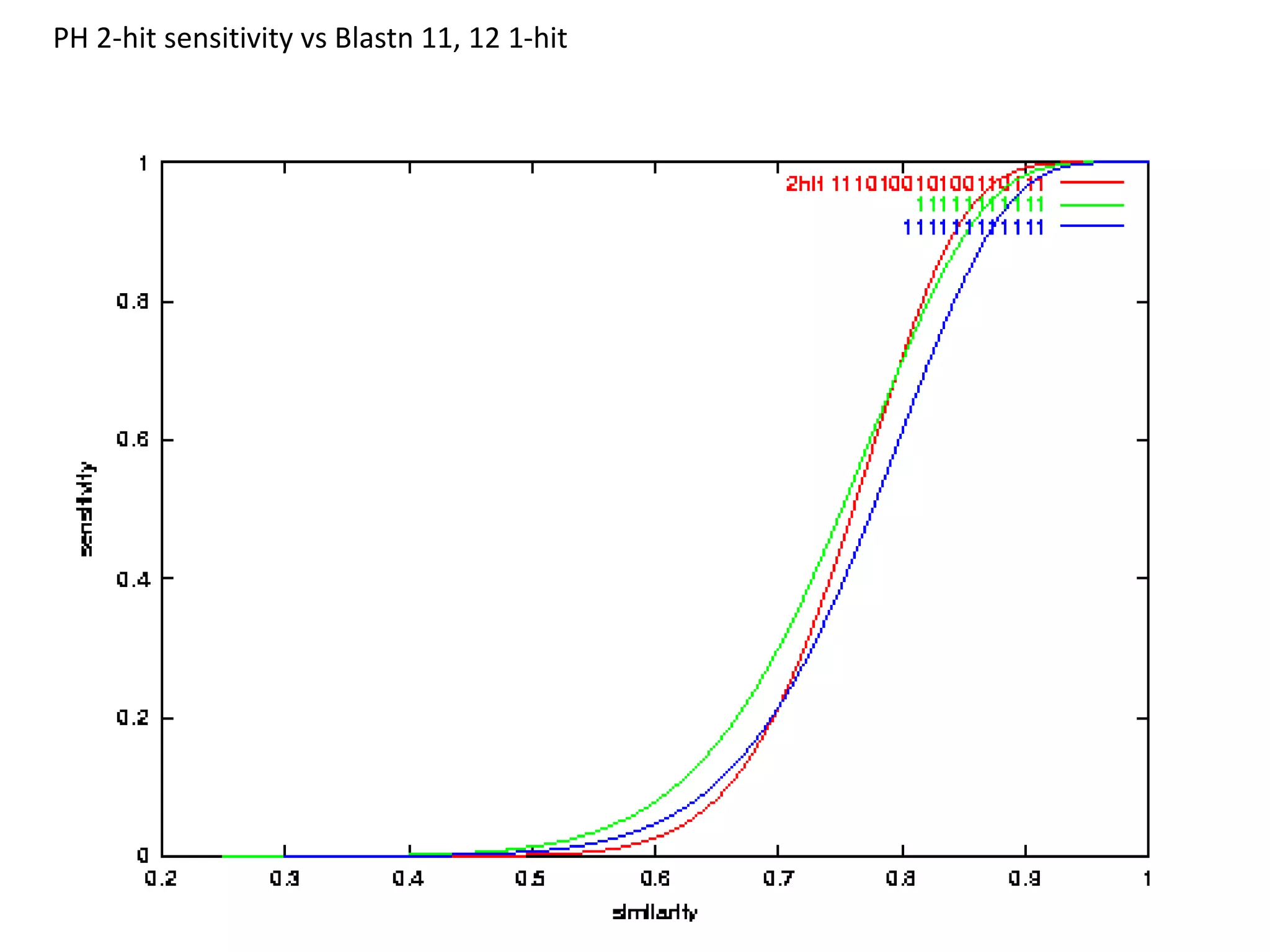
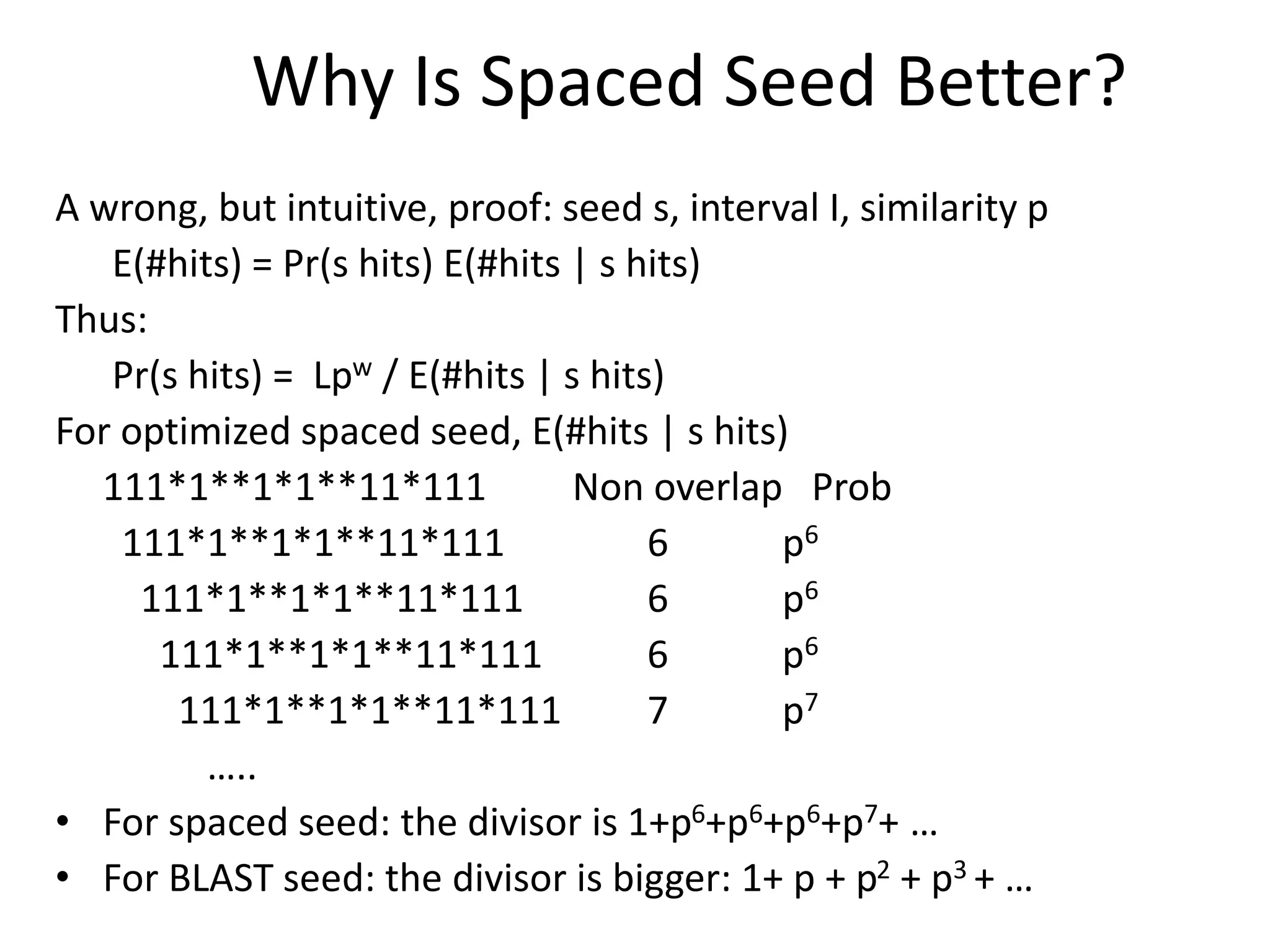
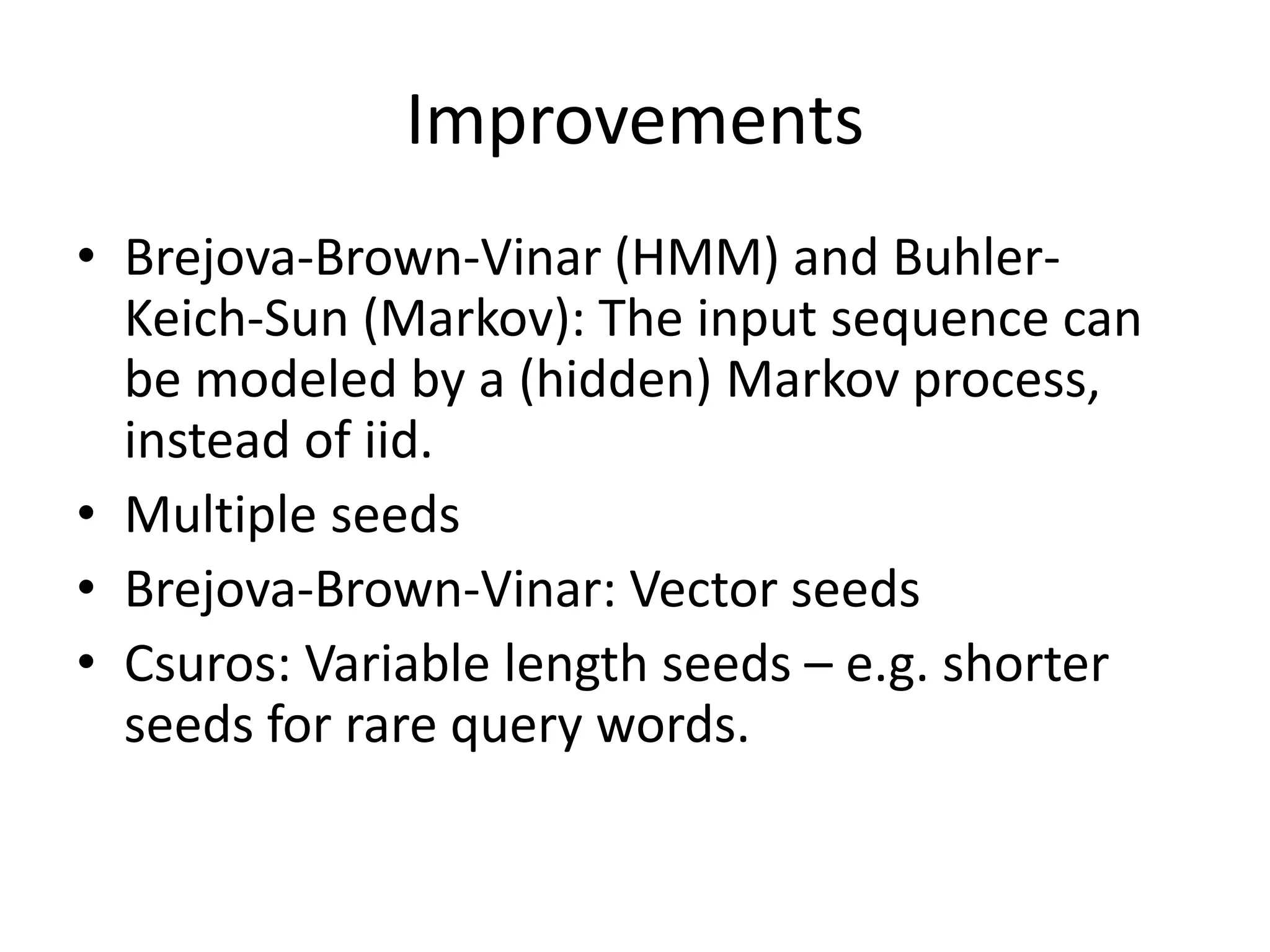
Spaced seeds are used for alignment and pattern matching in bioinformatics. They allow for some positions in the seed to be "don't care" positions represented by an asterisk, unlike BLAST seeds which require matches at every position. This simple change significantly increases the probability of hitting homologous regions while reducing spurious matches. Formally, the expected number of hits of a spaced seed within a region is lower than for a BLAST seed, but the probability of the seed hitting the region is higher, improving sensitivity. Multiple spaced seeds and hidden Markov models have also been used to improve alignment.