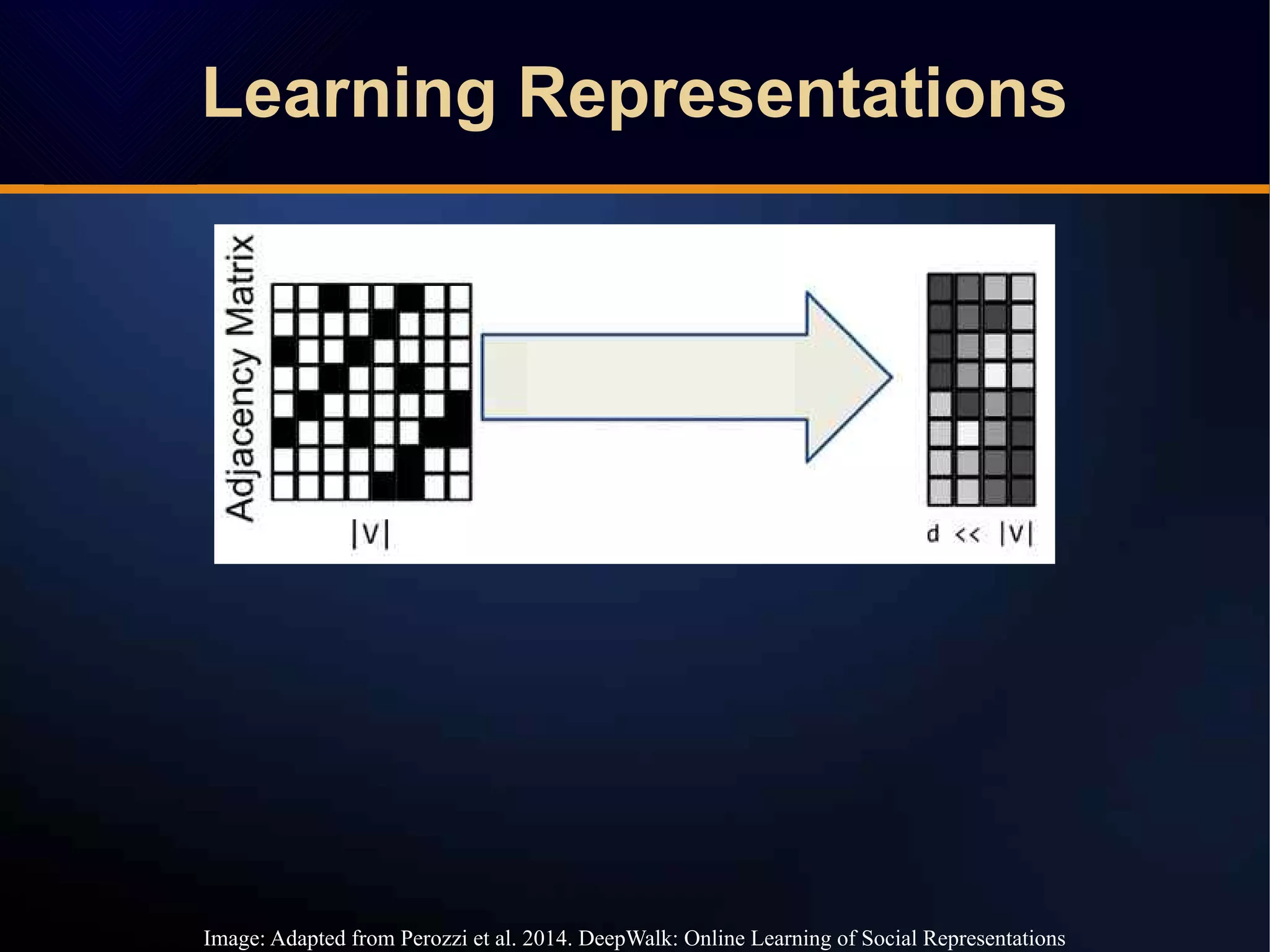
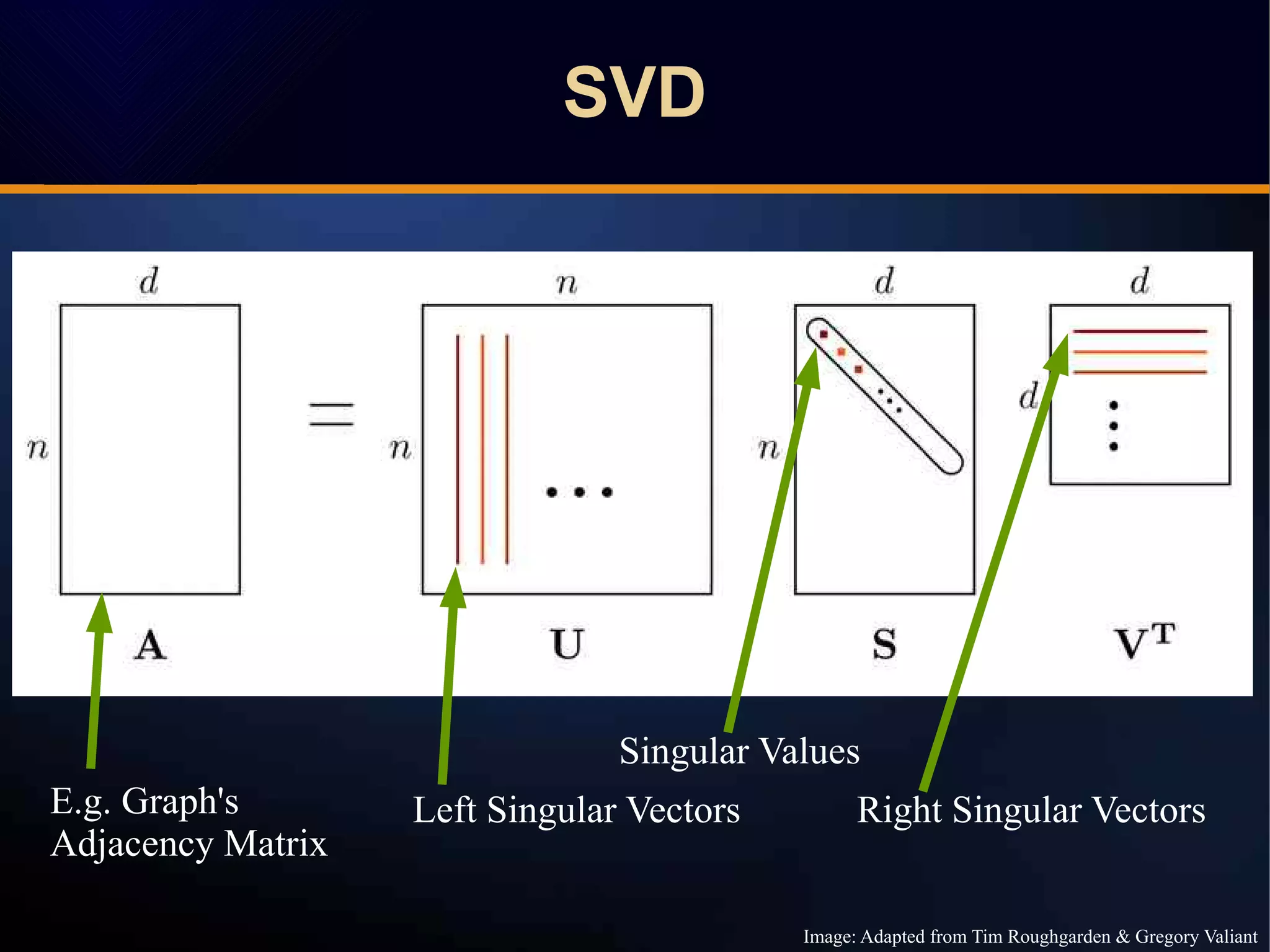
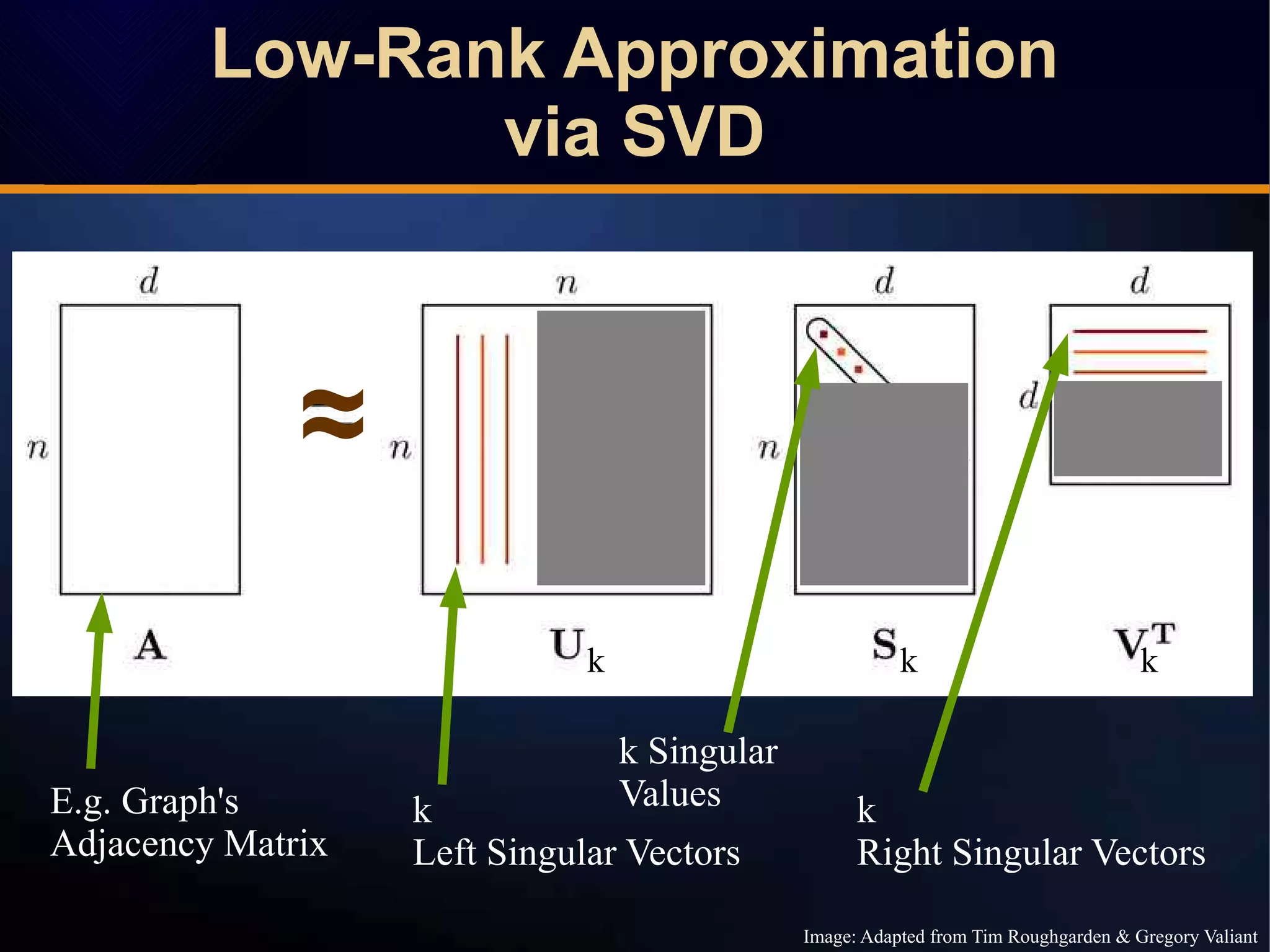
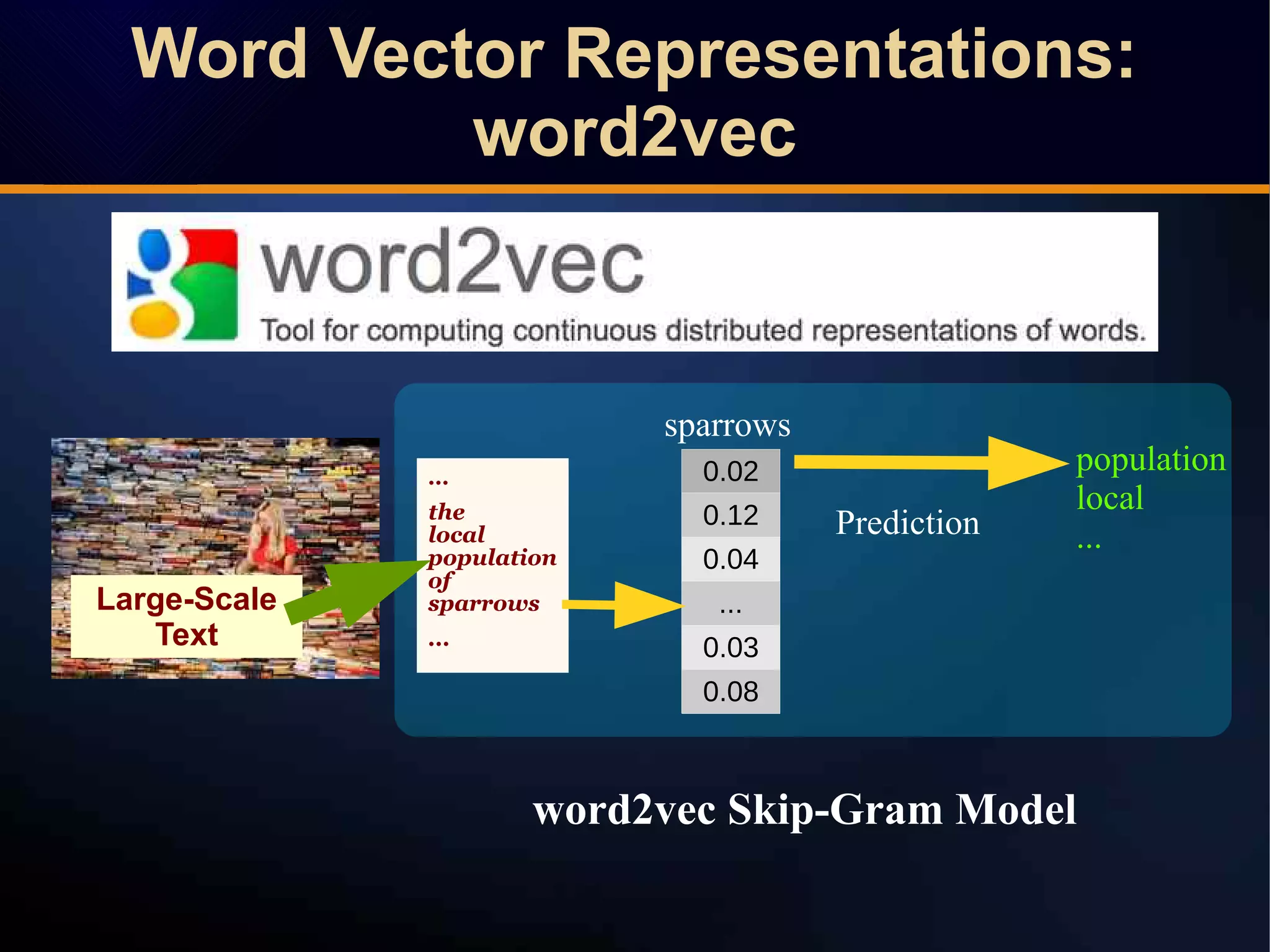
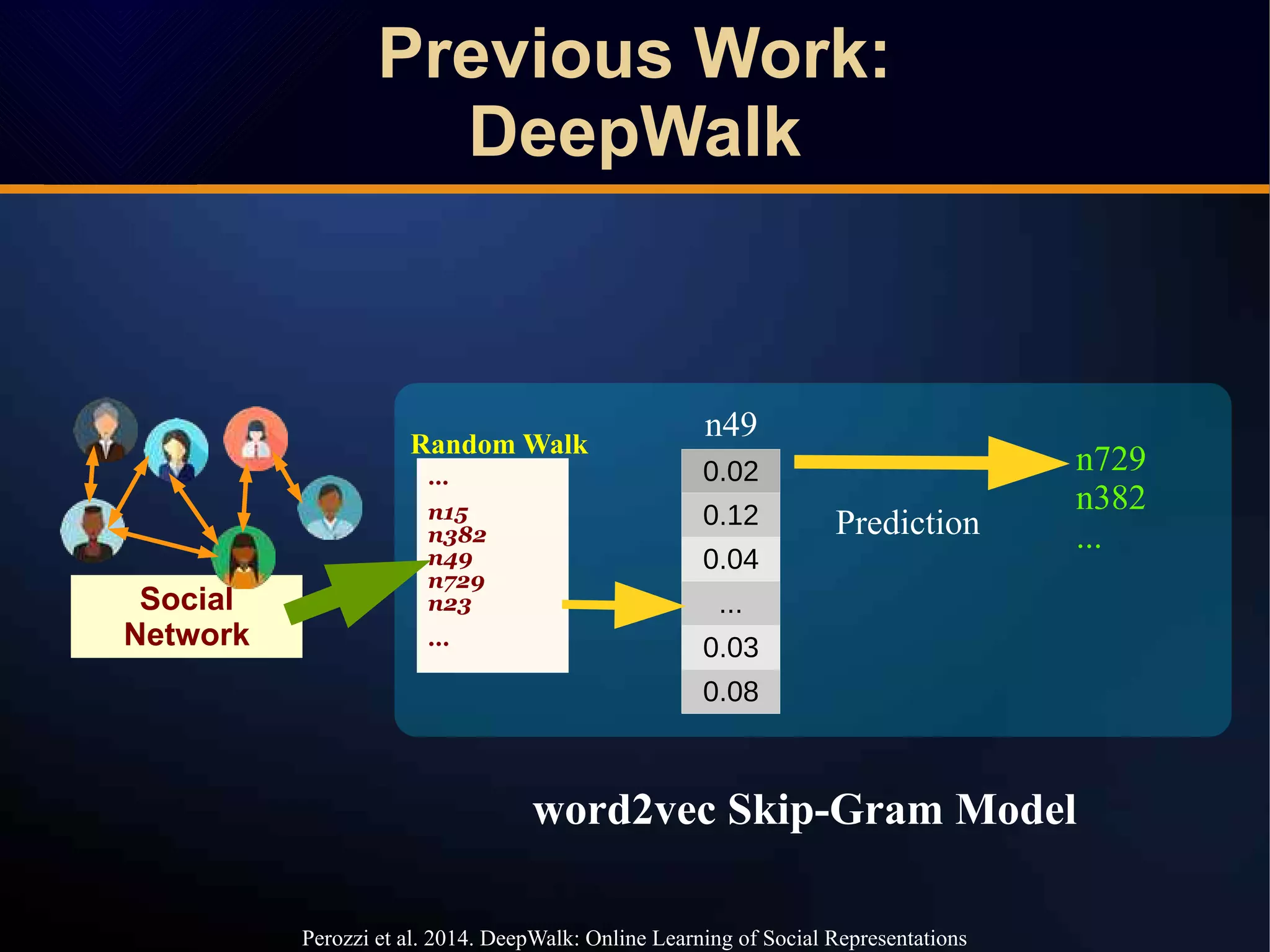
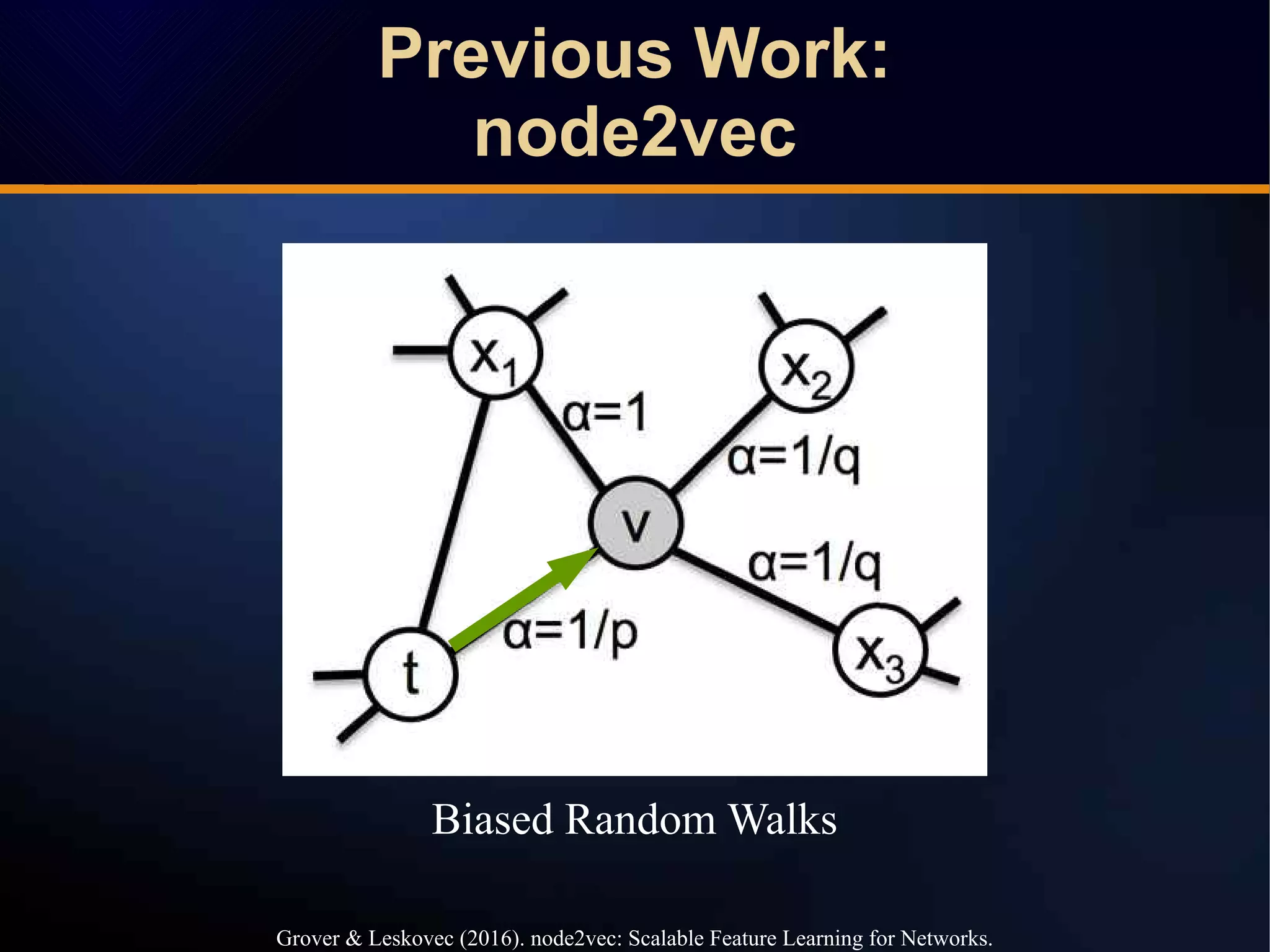
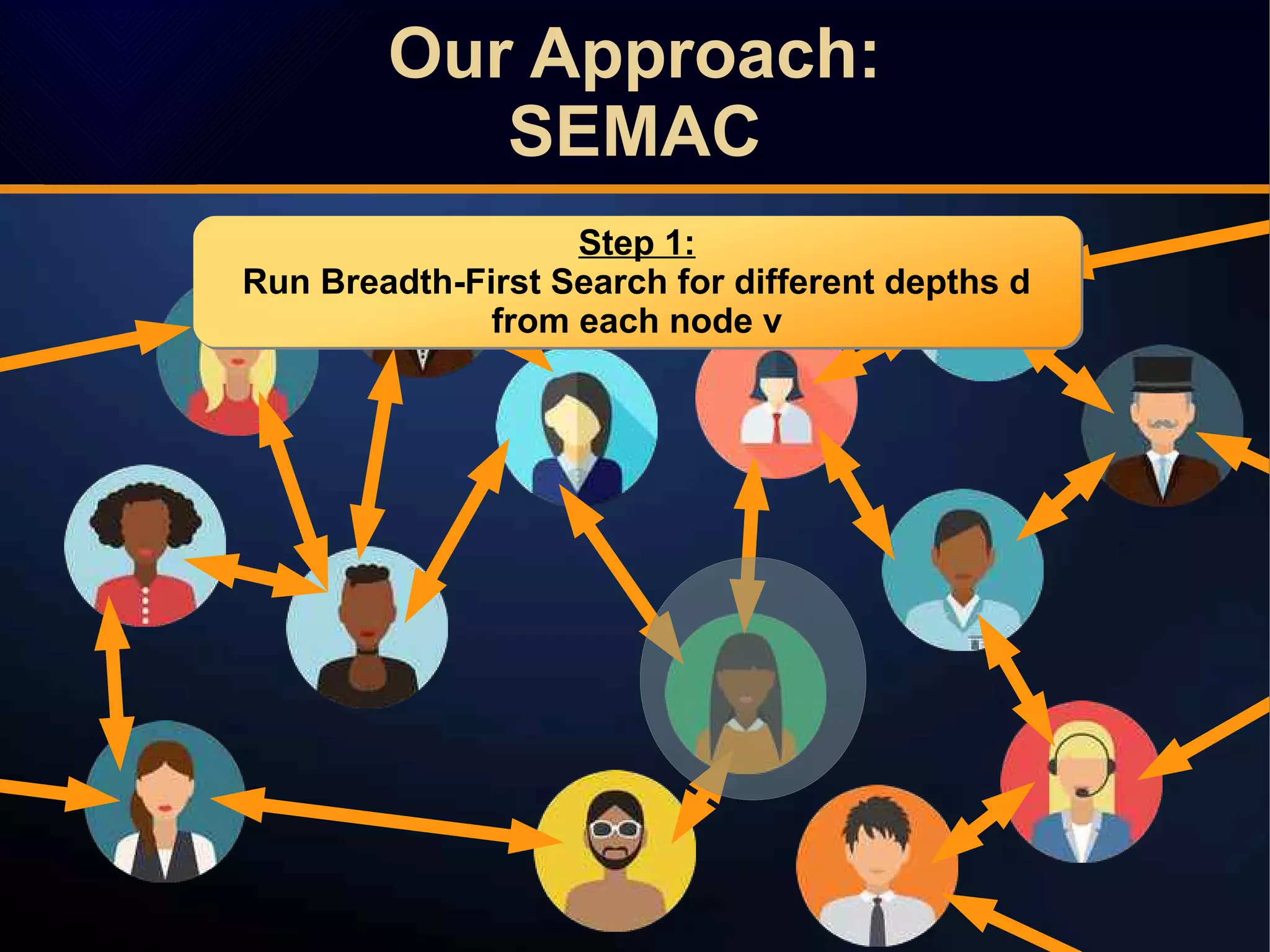
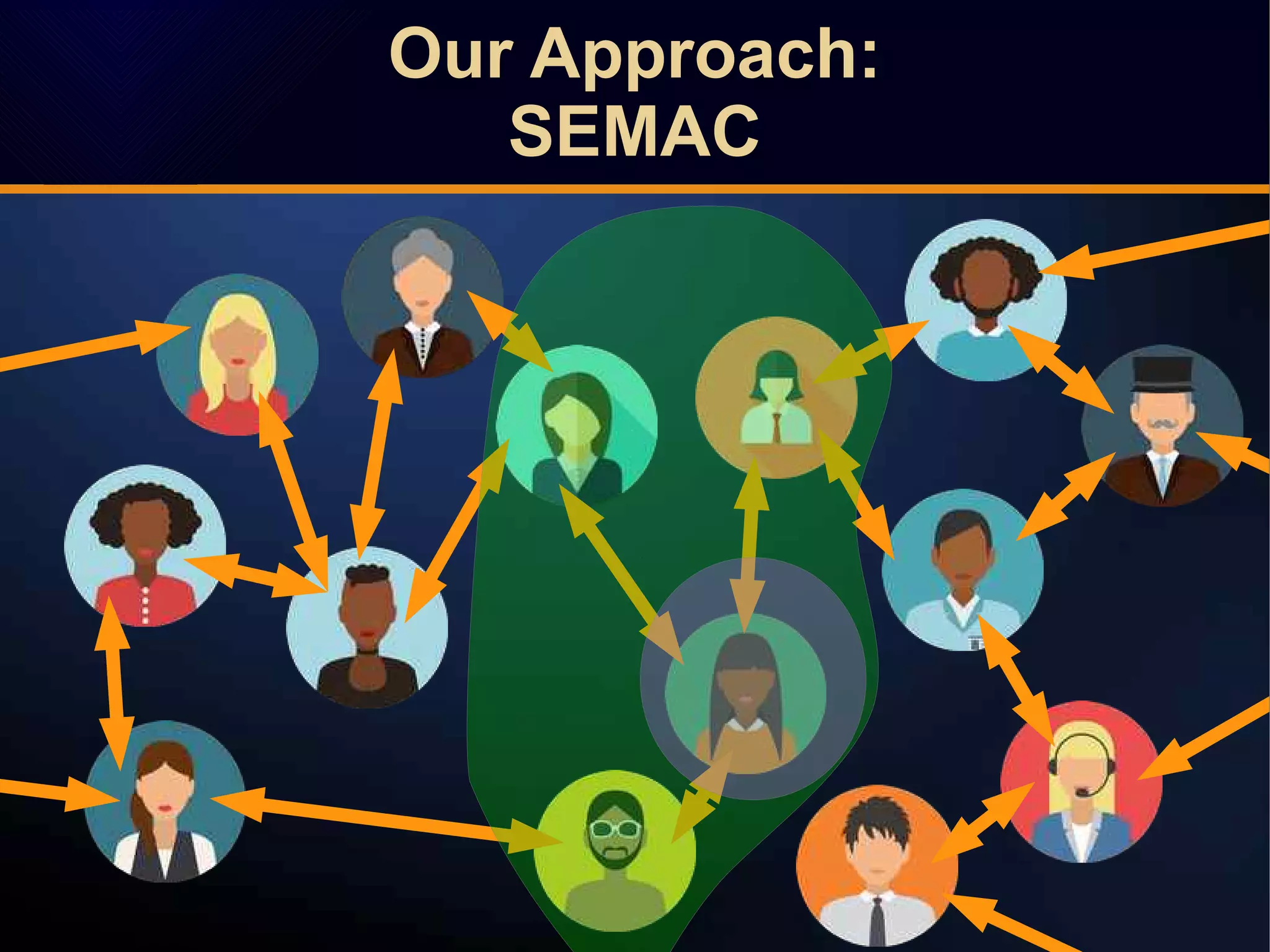
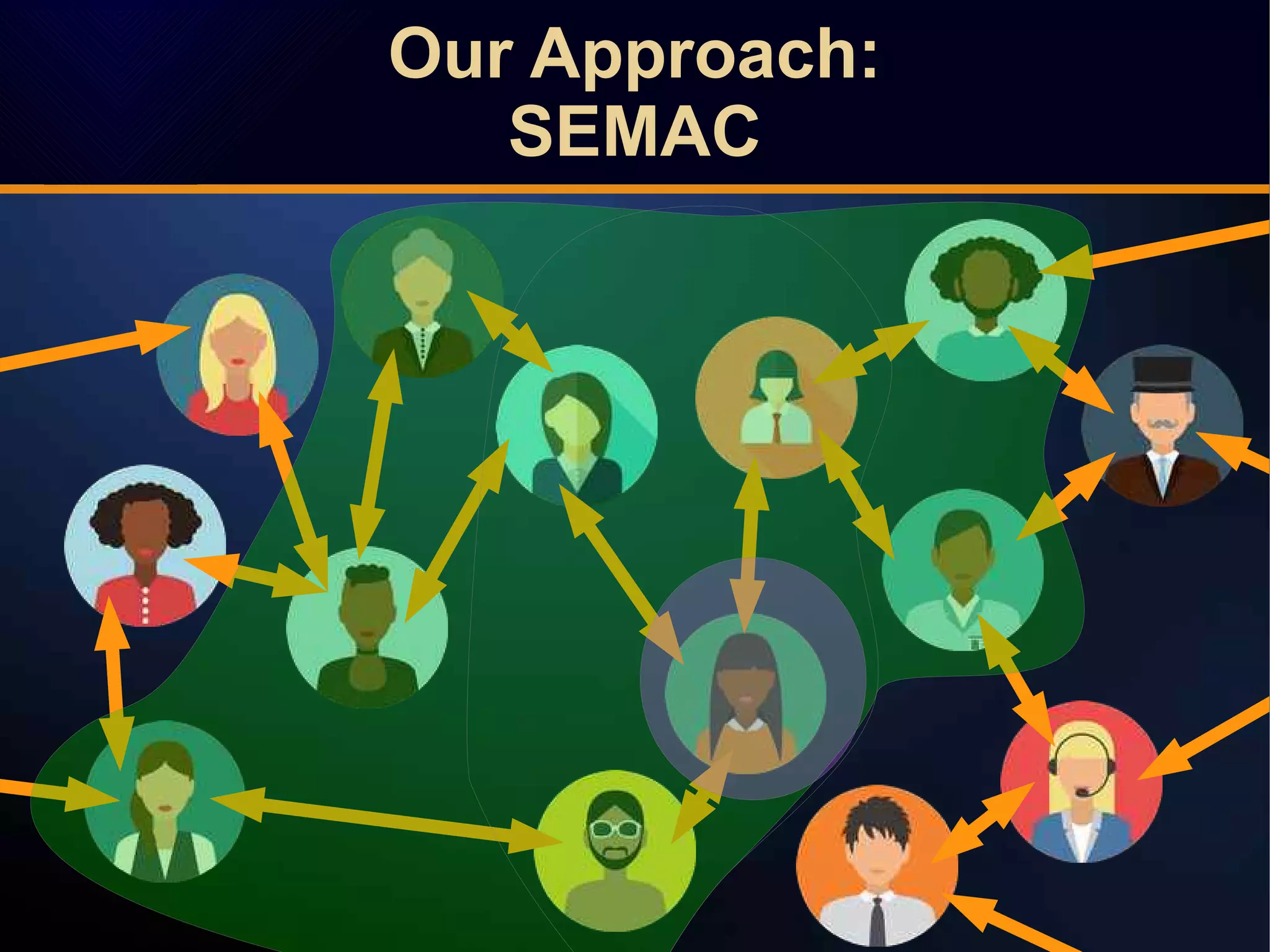
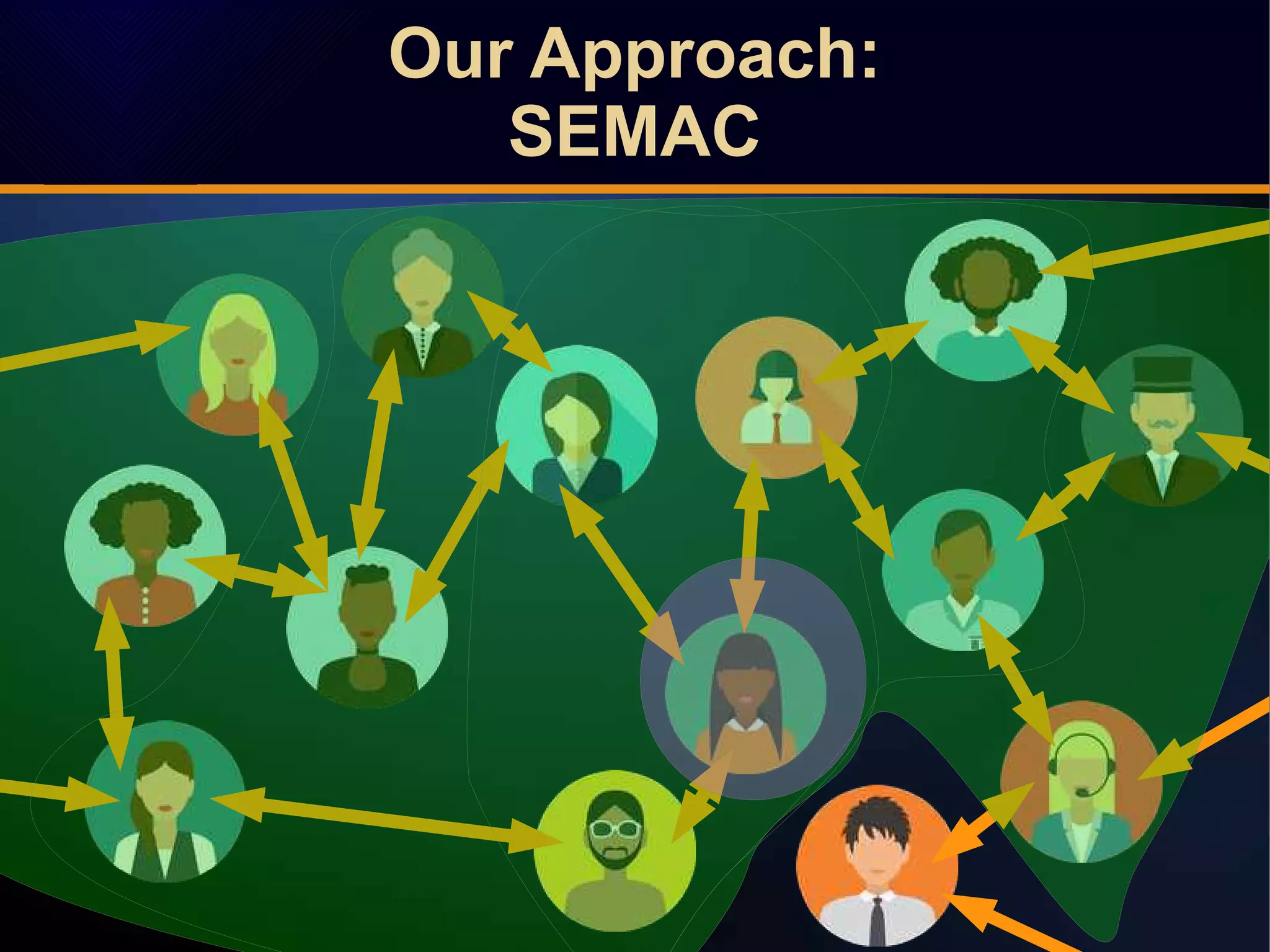
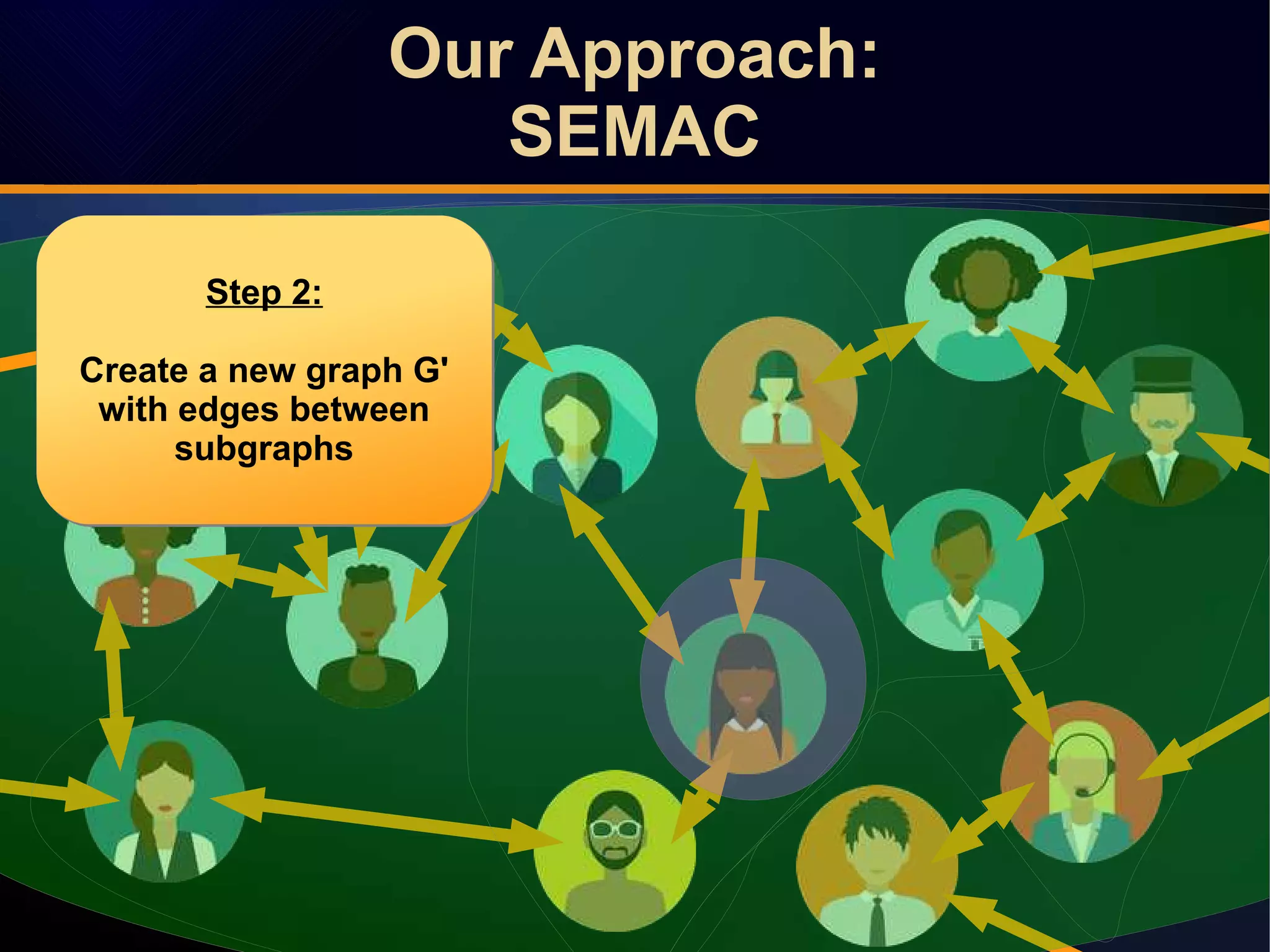
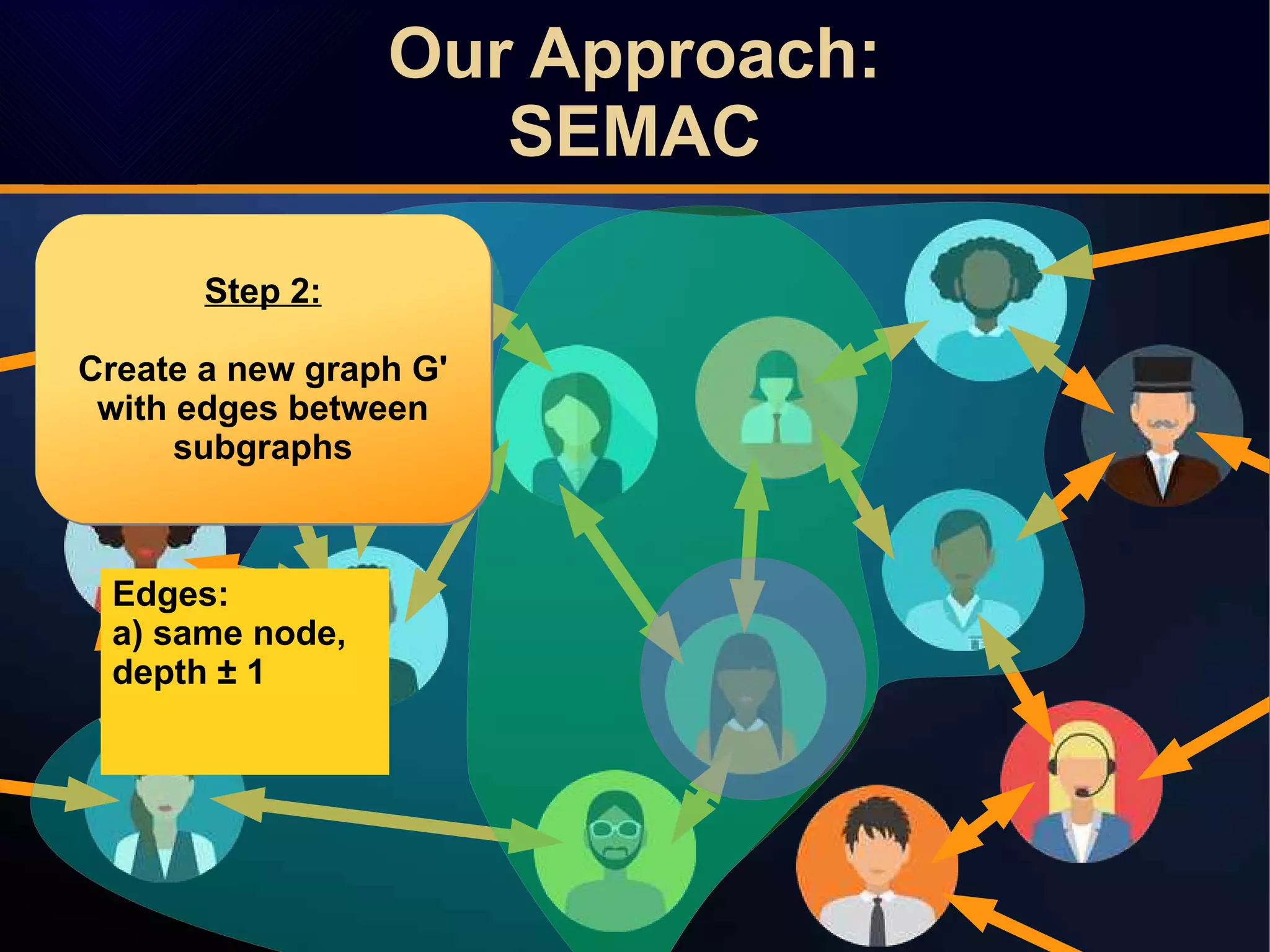
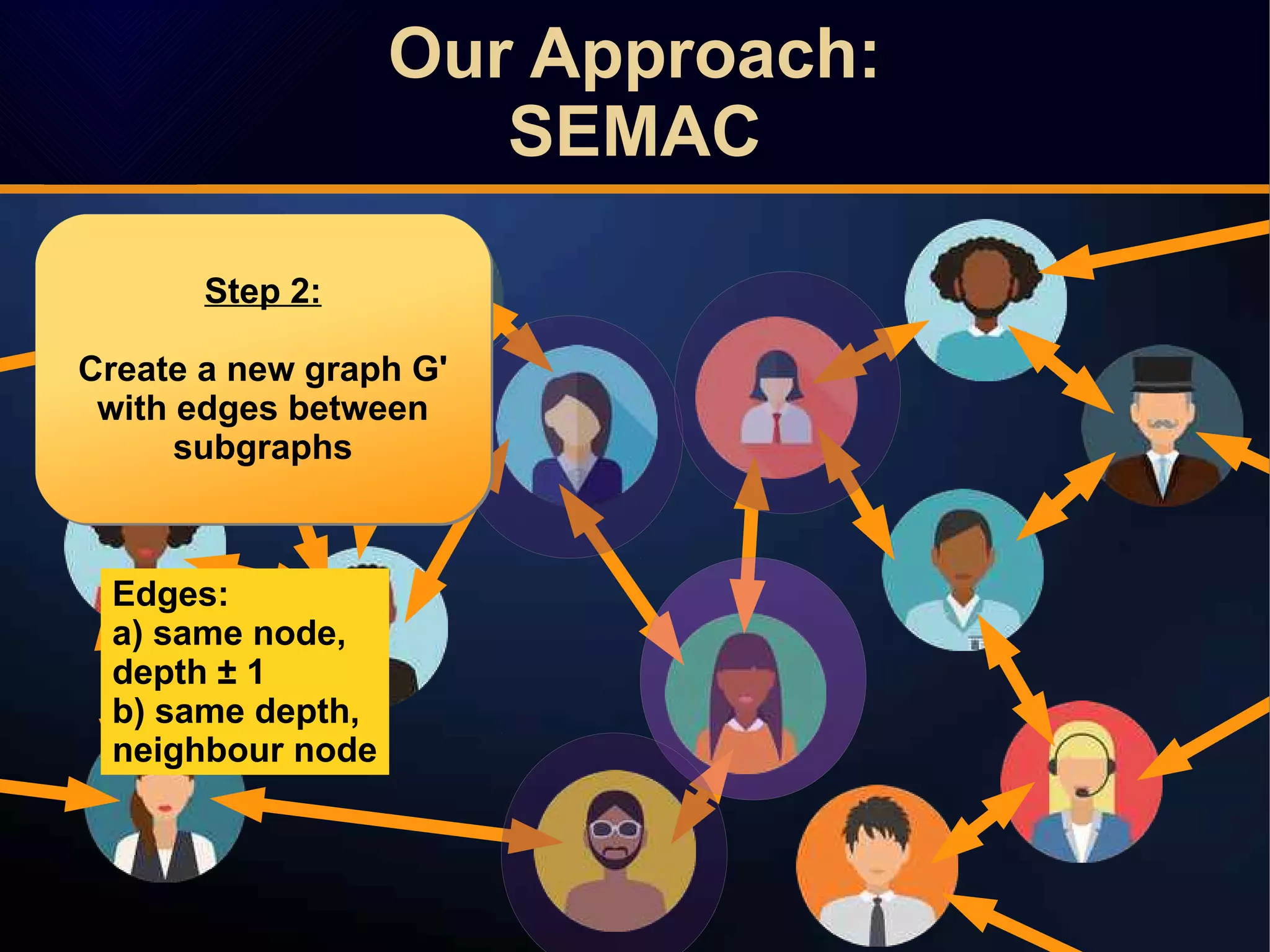
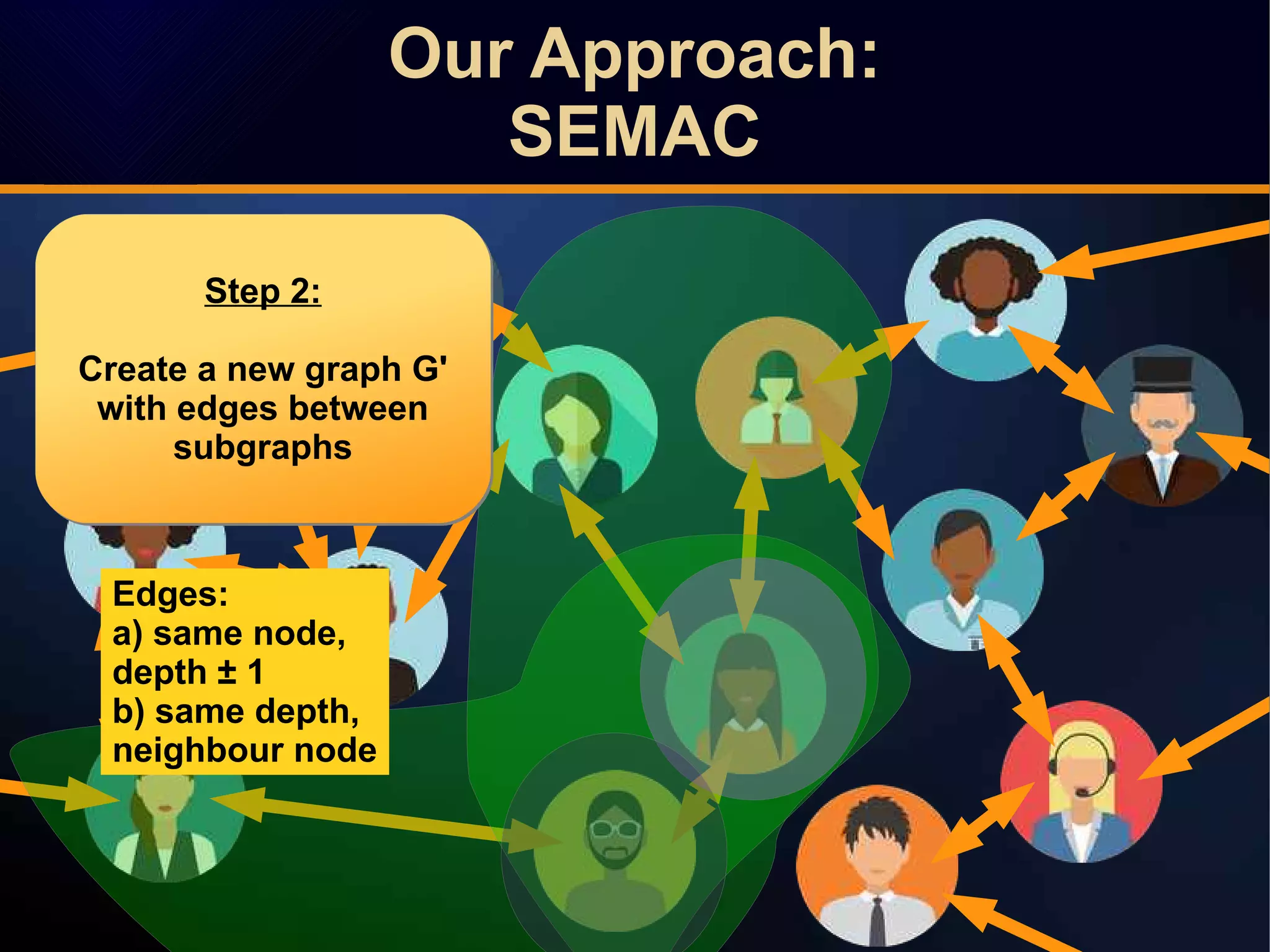
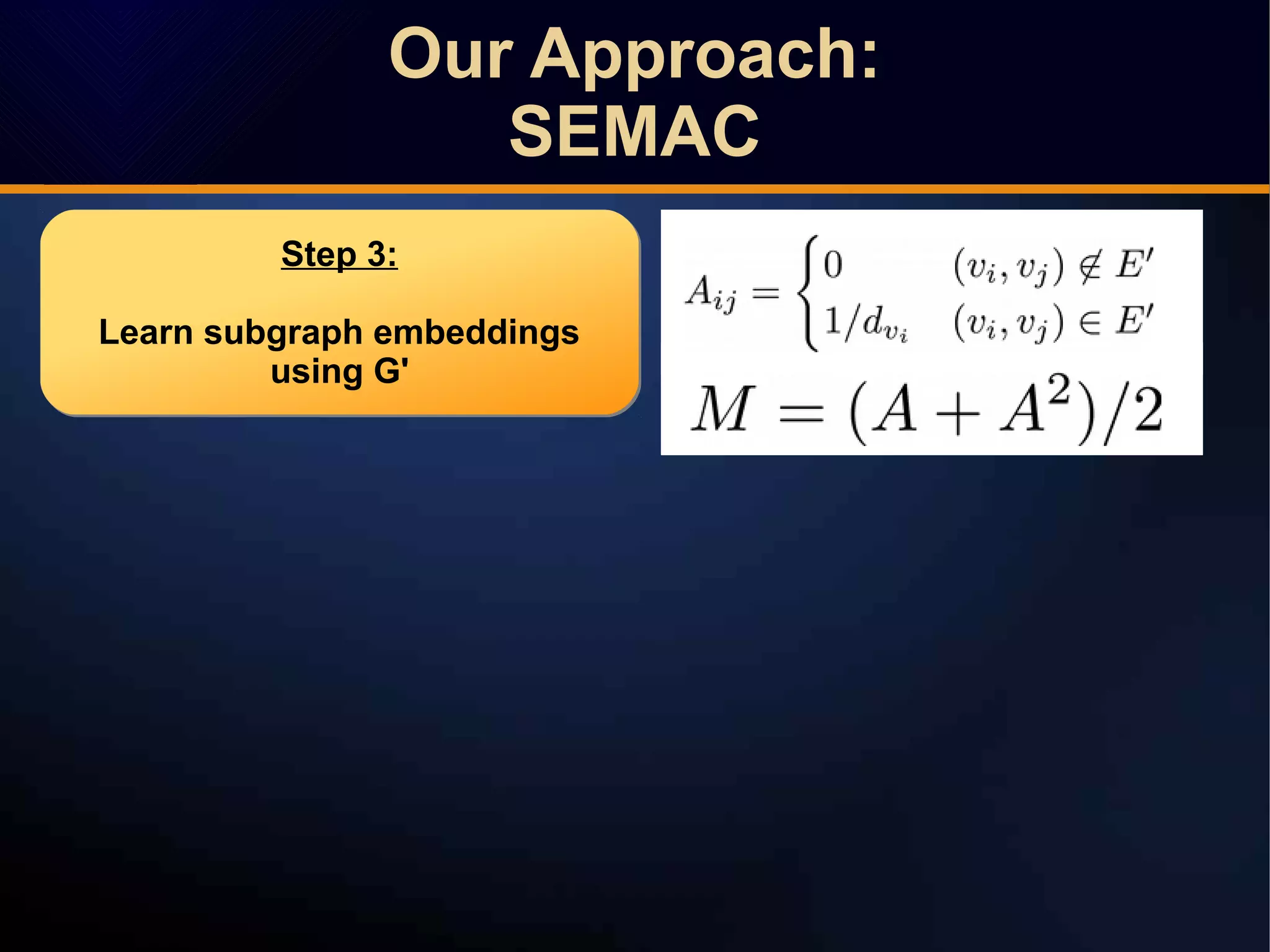
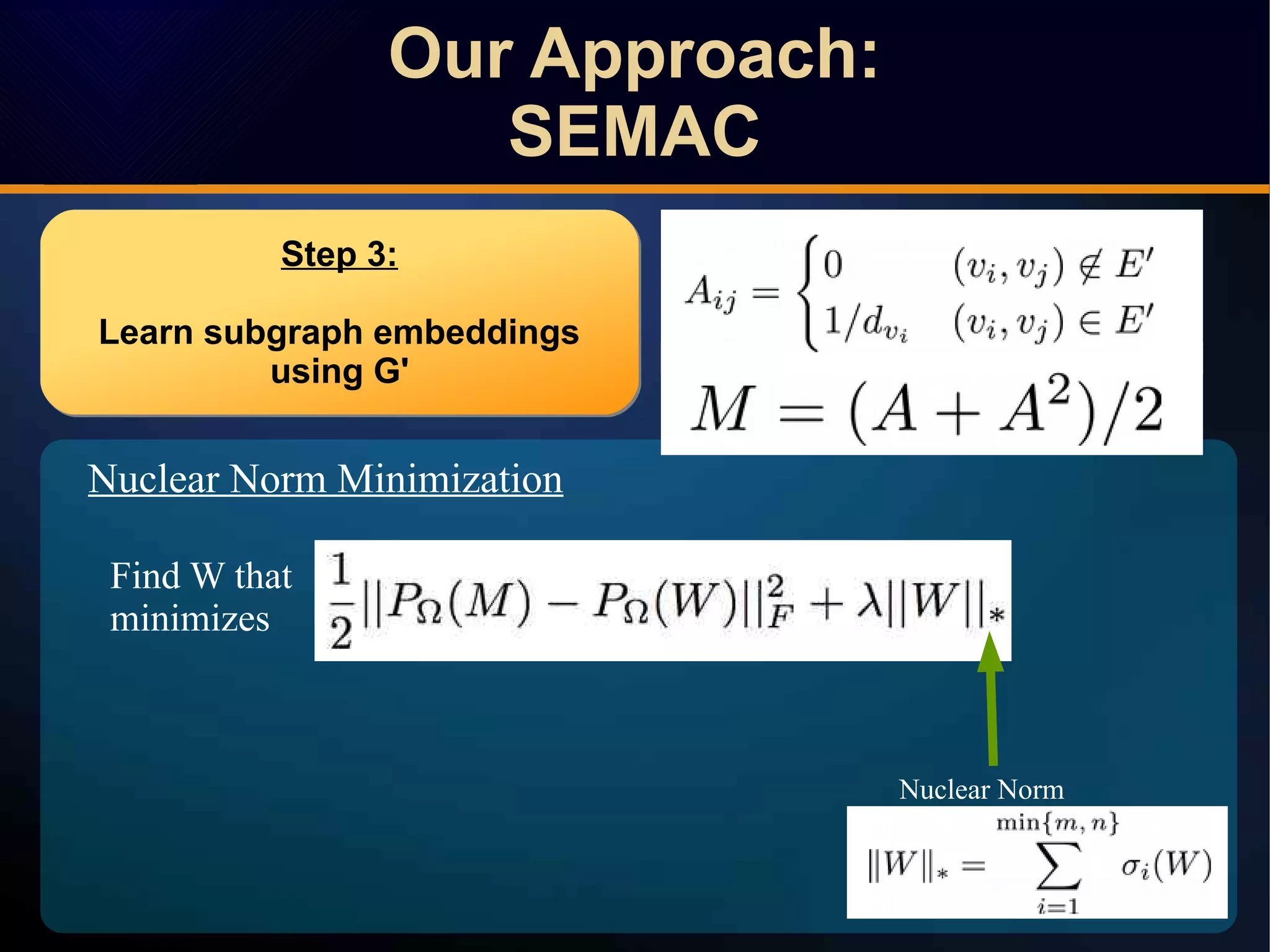
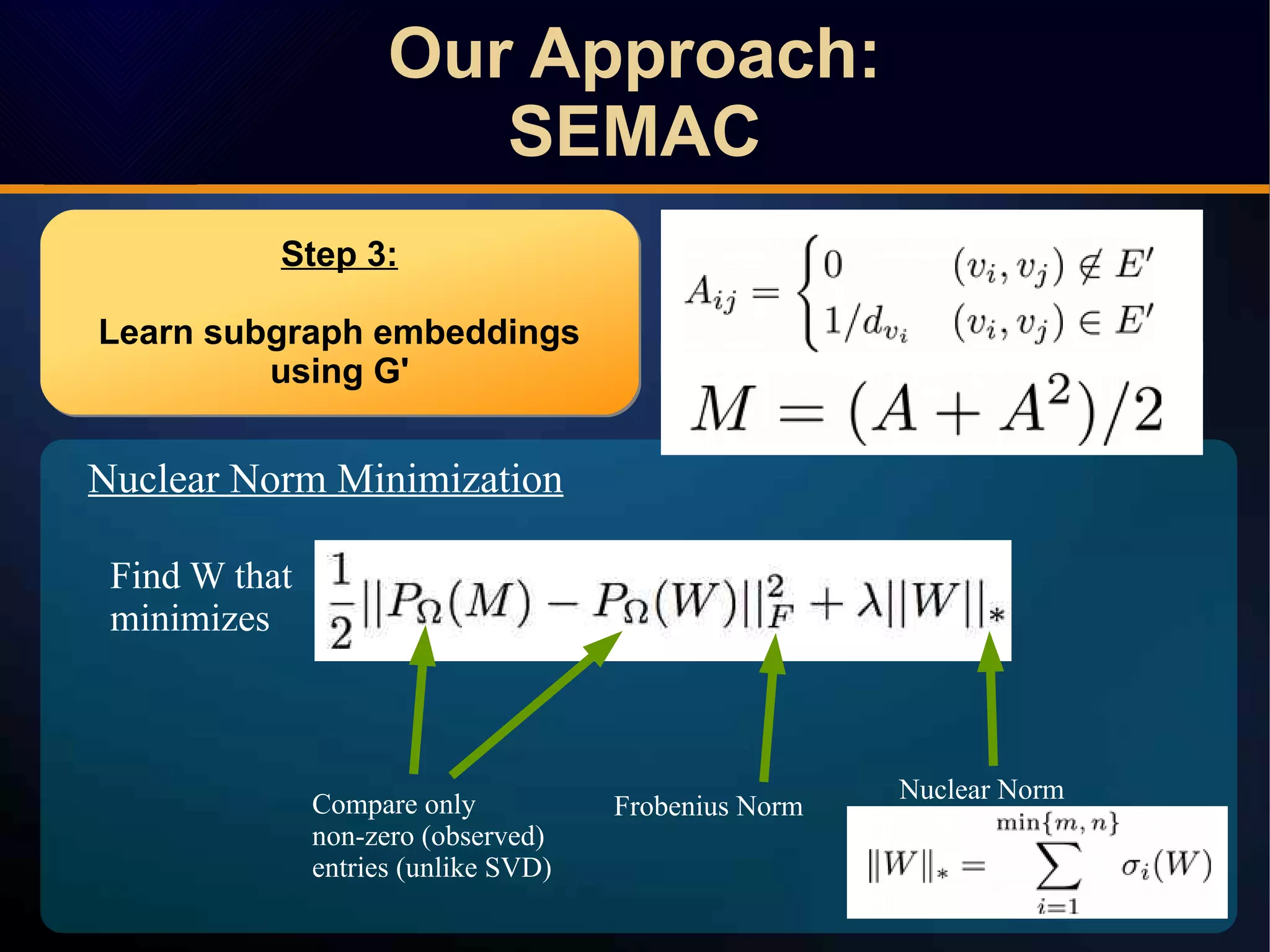
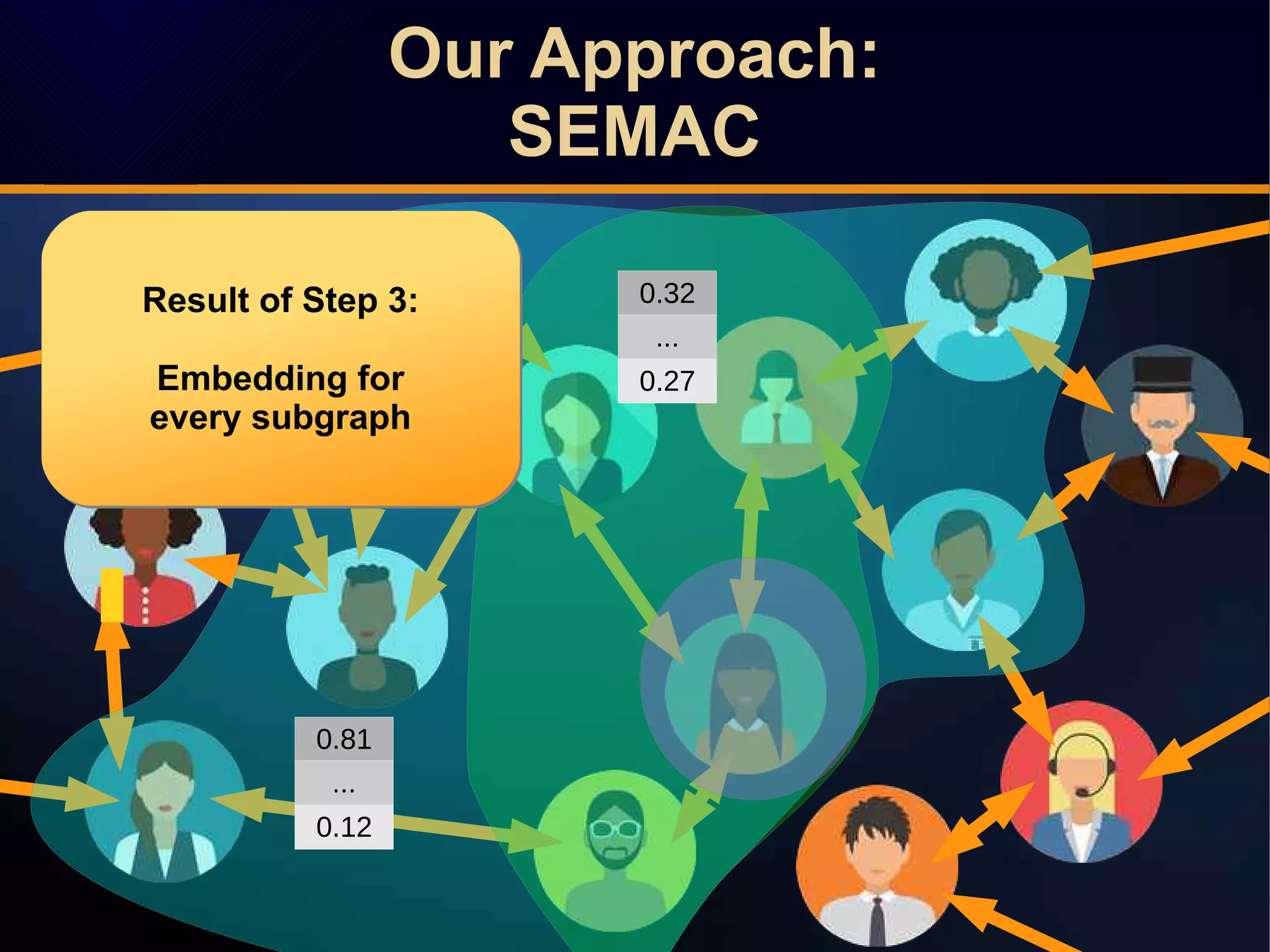
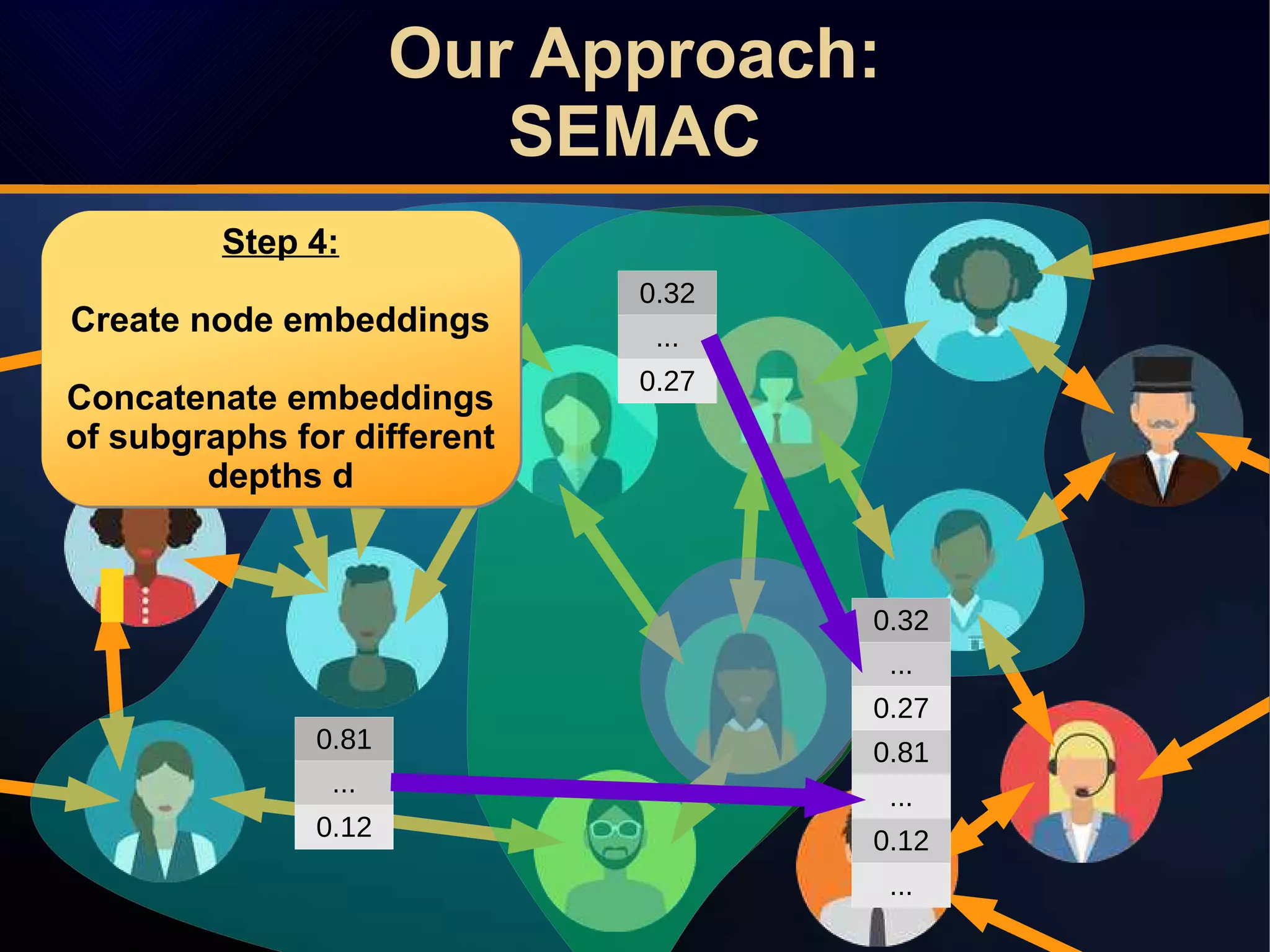
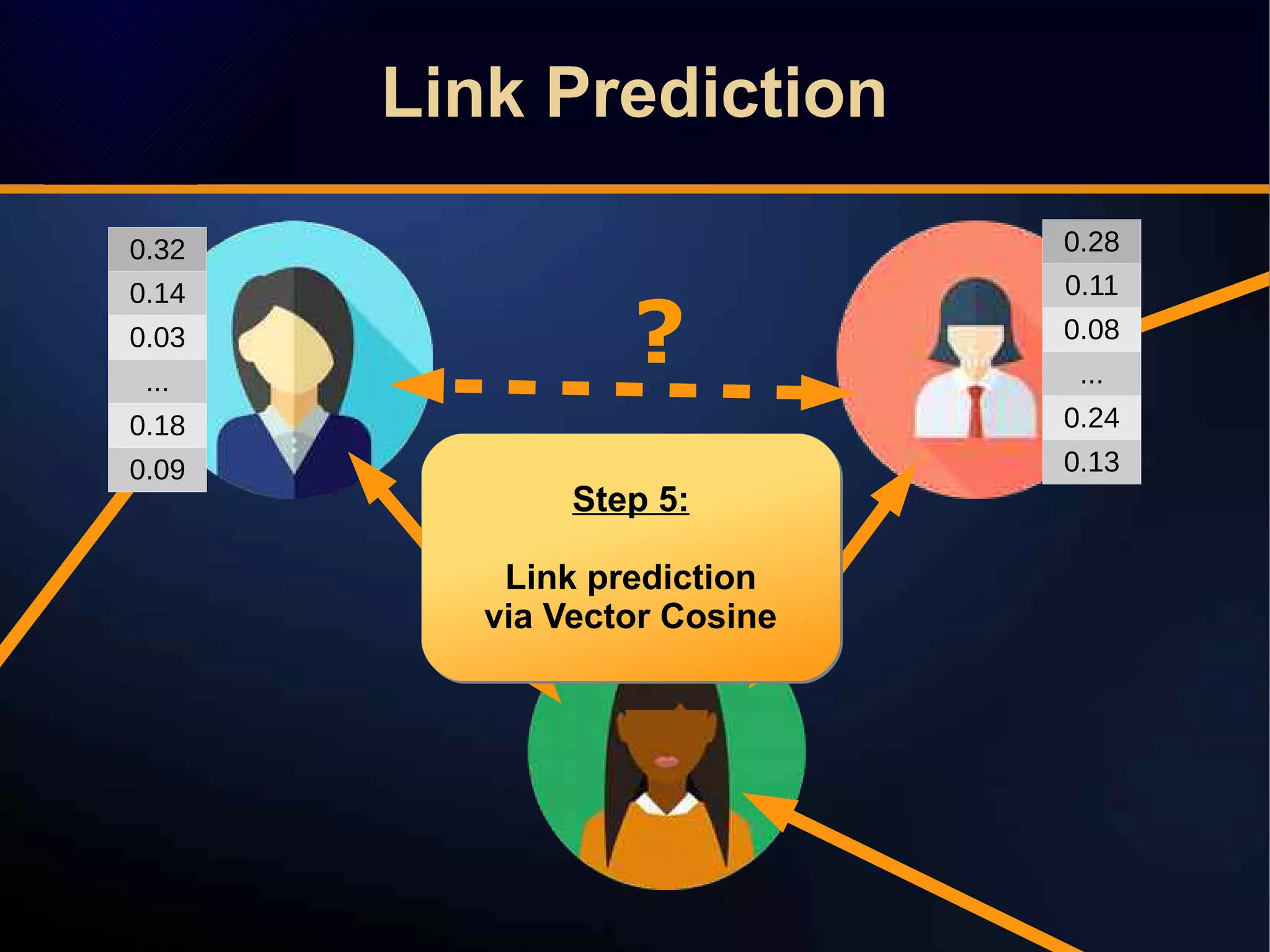
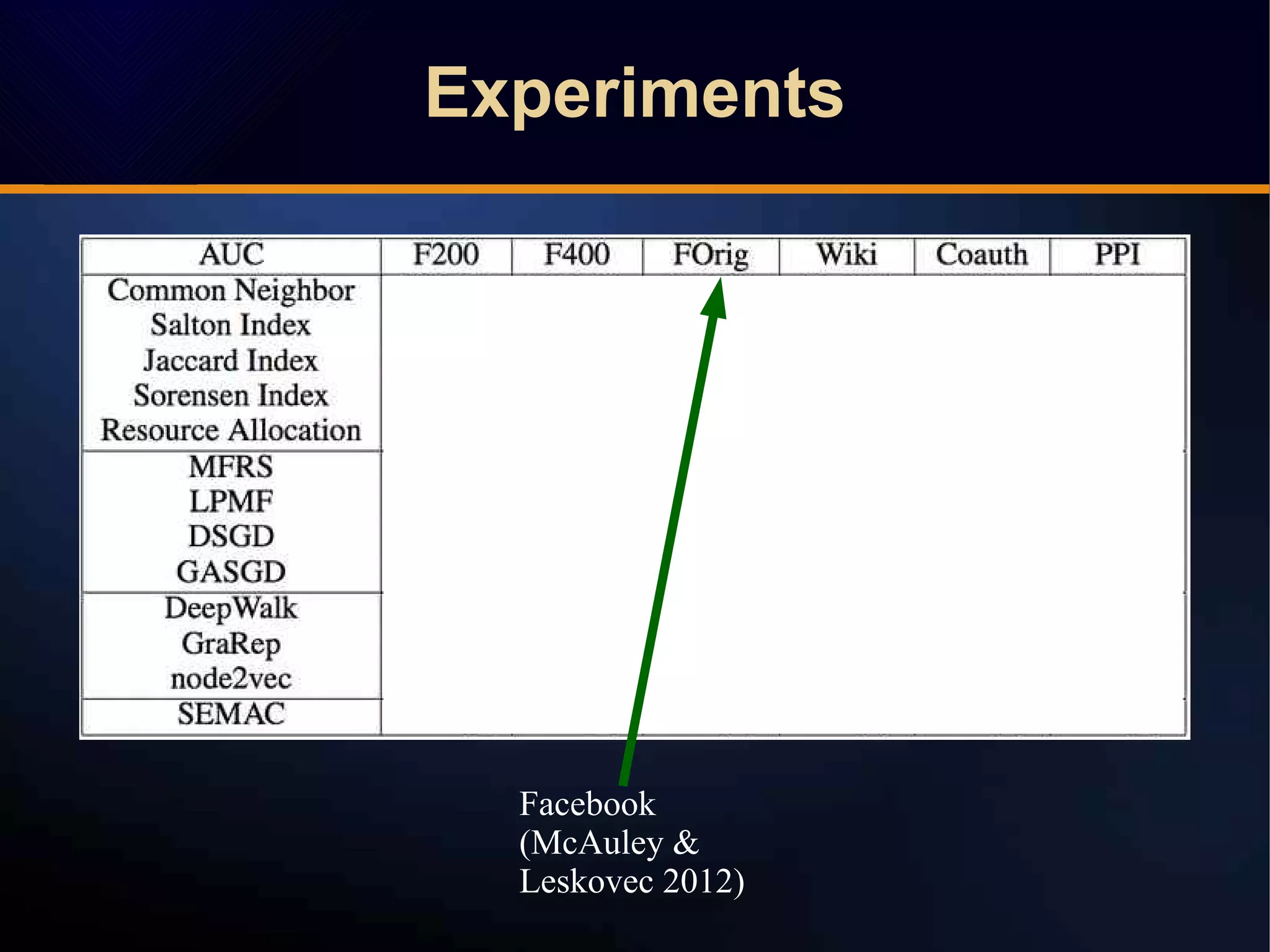
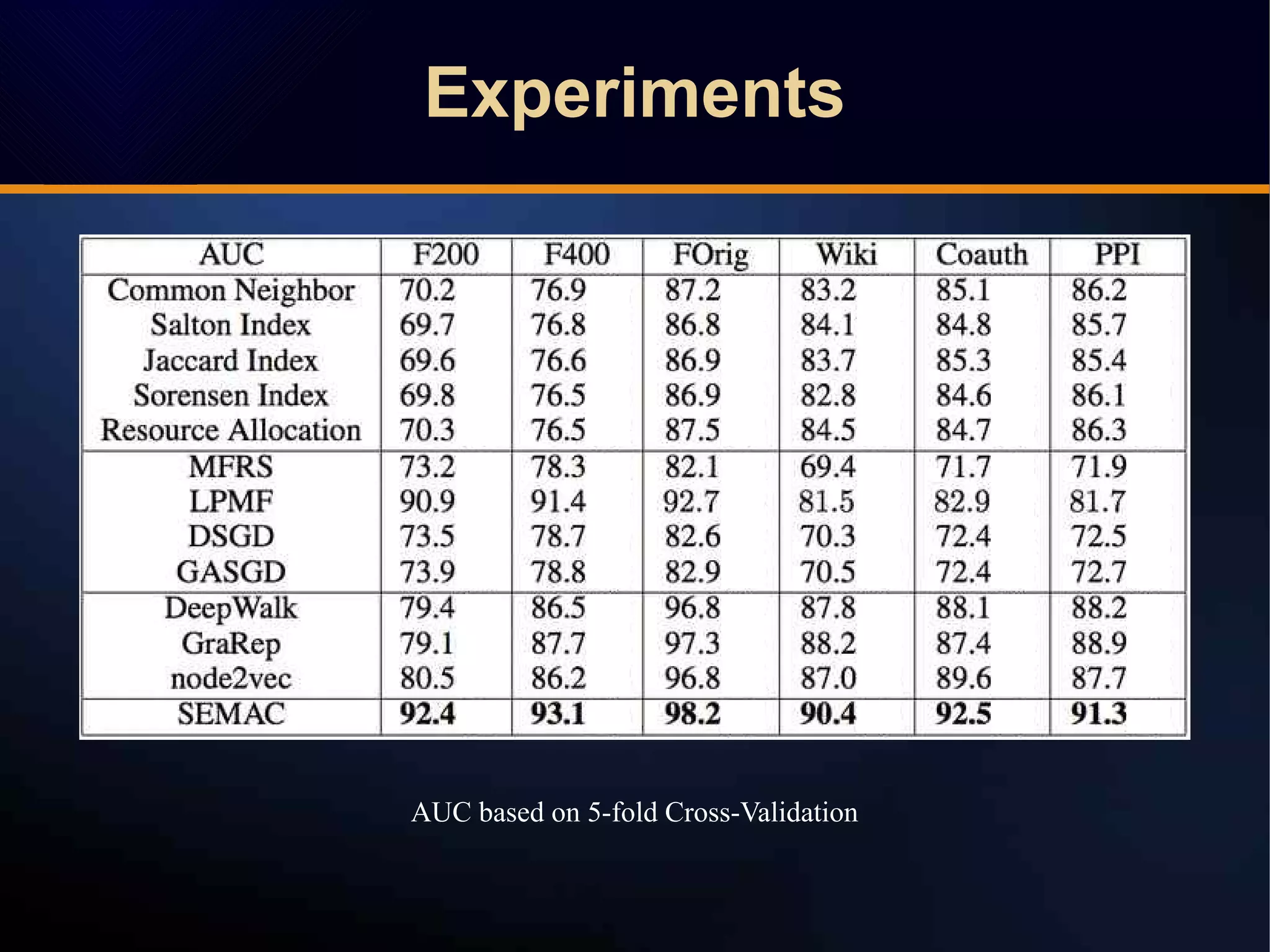
The document discusses a novel approach to link prediction through subgraph embedding and convex matrix completion, referred to as SEMAC. This method includes creating a new graph based on breadth-first search, learning subgraph embeddings, and using nuclear norm minimization for better accuracy in predicting connections. Results from various experiments on social networks, including Facebook and Wikipedia, indicate that SEMAC achieves state-of-the-art performance in link prediction tasks.