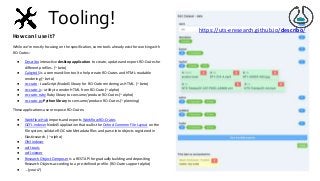
https://zbmed.github.io/damalos DaMaLOS 2020 workshop at ISWC 2020
The swings and roundabouts of a decade of fun and games with Research Objects
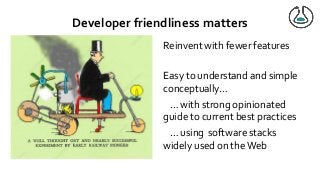
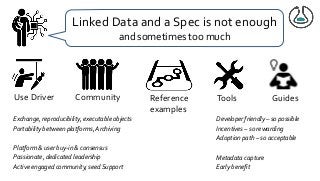
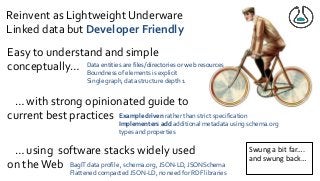
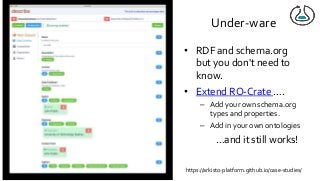
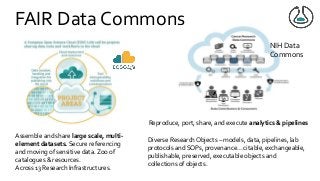
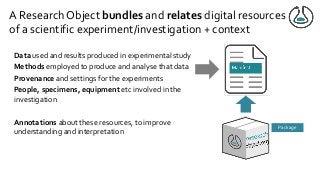
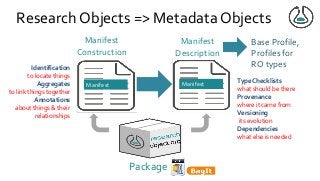
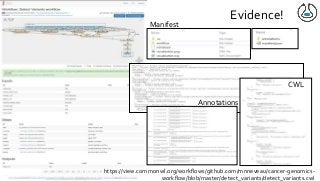
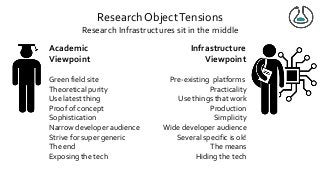
Over the past decade we have been working on the concept of Research Objects http://researchobject.org). We worked at the 20,000 feet level as a new form of FAIR+R scholarly communication and at the 5 feet level as a packaging approach for assembling all the elements of a research investigation, like the workflows, data and tools associated with a publication. Even in our earliest papers [1] we recognised that there was more to success than just using Linked Data. We swing from fancy RDF models and SHACL that specialists could use to simple RO-Crates and JSON-LD that mortal programmers could use. Success depends on a use case need, a community and tools which we found particularly in the world of computational workflows right from the outset. All along there have been politics and that has recently got even hotter. All fun and games!
[1] Sean Bechhofer, et al (2013) Why Linked Data is Not Enough for Scientists FGCS 29(2)599-611 https://doi.org/10.1016/j.future.2011.08.004 v






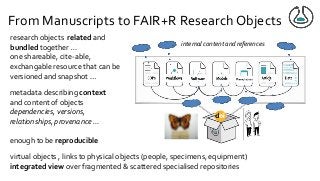
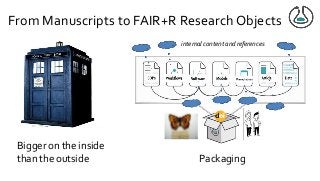
![From Manuscripts to Research Objects
“An article about computational science in a scientific publication is not the scholarship itself, it is
merely advertising of the scholarship.The actual scholarship is the complete software
development environment, [the complete data] and the complete set of instructions which
generated the figures.” David Donoho, “Wavelab and Reproducible Research,” 1995
research outcomes more than just
publications
data software, models, workflows, SOPs,
lab protocols are first class citizens of
scholarship
added information required to make
research FAIR and Reproducible (FAIR+R) …](https://image.slidesharecdn.com/damaloskeynotegoble2020-201102171100/85/The-swings-and-roundabouts-of-a-decade-of-fun-and-games-with-Research-Objects-8-320.jpg?cb=1604337116)

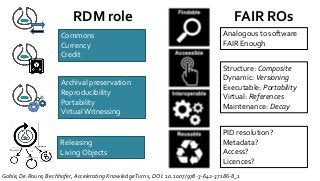


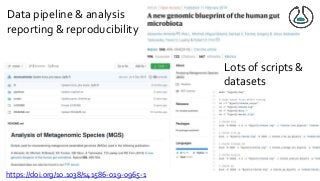
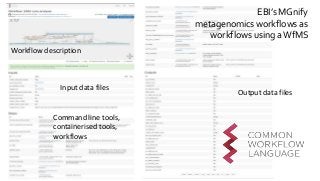
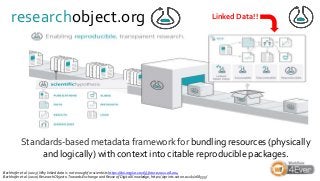



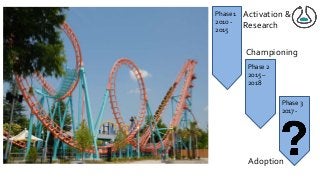


![Ladder Model of OSS Adoption
(adapted from Carbone P., Value Derived from Open
Source is a Function of Maturity Levels)
[FLOSS@Sycracuse]
"it's better, initially, to
make a small number of
users really love you than
a large number kind of
like you"
Paul Buchheit
paulbuchheit.blogspot.com](https://image.slidesharecdn.com/damaloskeynotegoble2020-201102171100/85/The-swings-and-roundabouts-of-a-decade-of-fun-and-games-with-Research-Objects-32-320.jpg?cb=1604337116)