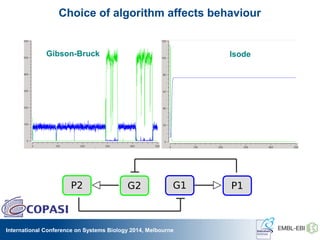
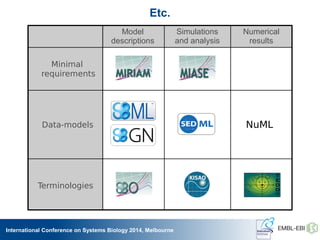
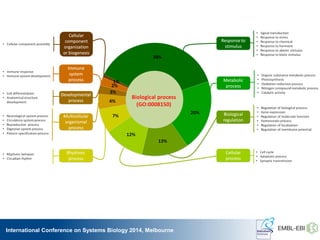
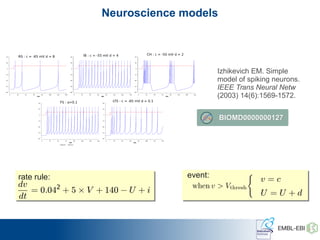
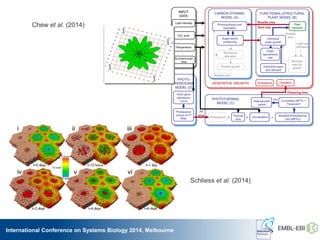
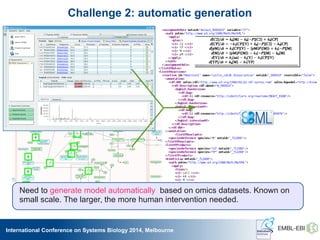
The International Conference on Systems Biology 2014 in Melbourne focused on the integration of standards and tools in systems biology, highlighting the evolution of computational models and the need for collaboration among researchers. Key discussions included the application of mathematical models in biology, the development of standard formats like SBML, and the challenges in modeling biological systems comprehensively. The conference emphasized the importance of modularity, reproducibility, and accessibility in biological modeling to facilitate experimentation and understanding.