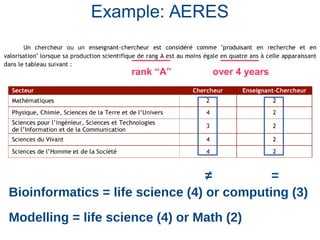
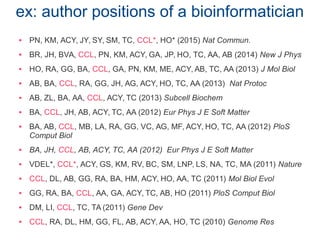
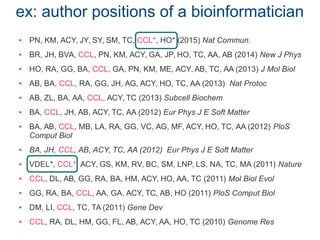
The document discusses the challenges of authorship and recognition in systems biology, emphasizing the underappreciation of bioinformatics and modeling contributions in scientific evaluations. It critiques the reliance on traditional publication metrics that favor certain author positions and journal prestige, advocating for more inclusive assessments of diverse research outputs. The document calls for policy changes to recognize varied contributions within the scientific community, including datasets, technical specifications, and software tools.