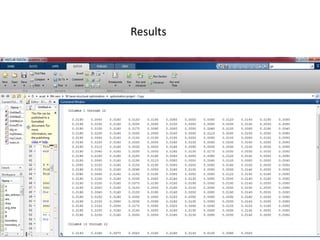
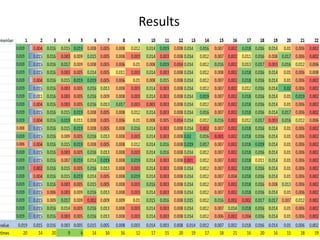
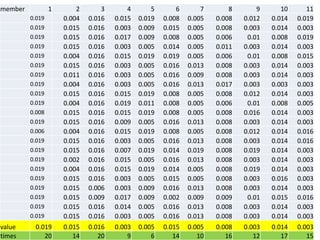
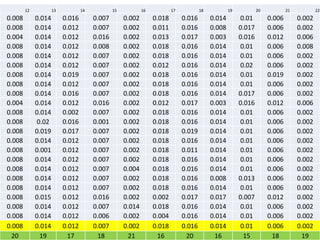
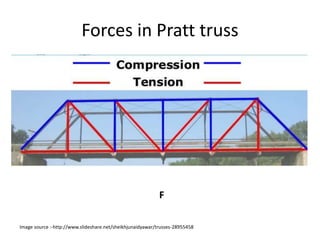
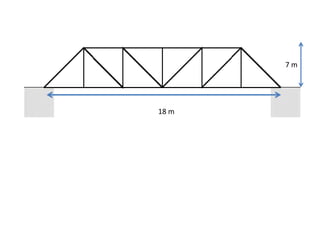
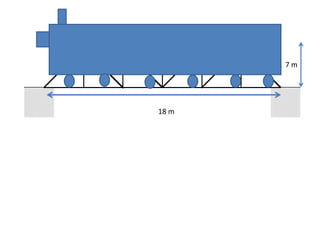
The document describes optimizing the design of a Pratt truss bridge using a genetic algorithm. It presents the objective of minimizing the total cross-sectional area of the truss members subject to constraints on member size and displacement. The genetic algorithm is applied over multiple generations using selection, crossover and mutation to iteratively evolve solutions toward an optimal design. The results show the optimized cross-sectional area in cm^2 for each of the 22 truss members.



![clc; clear all;
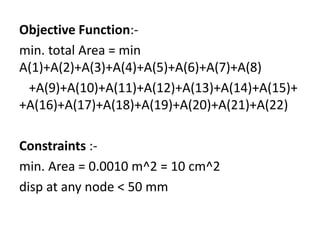
% initial cromosome having area in the range 0.002 to 0.02 m^2-
C = 2e-3*[1 2 4 10 3 9 10 7 1 5 1 1 2 4 10 3 9 10 7 1 5 2;
9 3 7 9 4 7 5 2 4 9 1 9 3 7 9 4 7 5 2 4 9 3;
7 3 4 1 1 7 3 2 4 9 8 7 3 4 1 1 7 3 2 4 9 4;
1 5 7 9 4 9 5 2 4 9 6 1 5 7 9 4 9 5 2 4 9 5;
4 10 6 2 2 2 3 2 4 1 9 4 10 6 2 2 2 3 2 4 1 6;
9 3 5 9 5 7 5 1 4 9 1 9 3 5 9 5 7 5 1 4 9 7;
9 9 4 1 4 3 1 2 4 9 8 9 9 4 1 4 3 1 2 4 9 8;
1 3 7 9 1 7 5 5 4 4 1 1 3 7 9 1 7 5 5 4 4 9;
9 8 3 1 2 4 2 2 4 9 6 9 8 3 1 2 4 2 2 4 9 10;
2 3 7 9 3 7 5 9 4 3 1 2 3 7 9 3 7 5 9 4 3 1;
8 7 2 10 4 5 3 10 4 7 4 8 7 2 10 4 5 3 10 4 7 2;
1 2 4 10 3 9 10 7 1 5 1 1 2 4 10 3 9 10 7 1 5 3;
9 3 7 9 4 7 5 2 4 9 1 9 3 7 9 4 7 5 2 4 9 4;
7 3 4 1 1 7 3 2 4 9 8 7 3 4 1 1 7 3 2 4 9 5;
1 5 7 9 4 9 5 2 4 9 6 1 5 7 9 4 9 5 2 4 9 6;
4 10 6 2 2 2 3 2 4 1 9 4 10 6 2 2 2 3 2 4 1 7;
9 3 5 9 5 7 5 1 4 9 1 9 3 5 9 5 7 5 1 4 9 8;
9 9 4 1 4 3 1 2 4 9 8 9 9 4 1 4 3 1 2 4 9 9;
1 3 7 9 1 7 5 5 4 4 1 1 3 7 9 1 7 5 5 4 4 10;
9 8 3 1 2 4 2 2 4 9 6 9 8 3 1 2 4 2 2 4 9 1;
2 3 7 9 3 7 5 9 4 3 1 2 3 7 9 3 7 5 9 4 3 2;
2 3 7 9 3 7 5 9 4 3 1 2 3 7 9 3 7 5 9 4 3 3];](https://image.slidesharecdn.com/pratttrussoptimizationusing-161116063529/85/Pratt-truss-optimization-using-9-320.jpg)




![%% Truss code
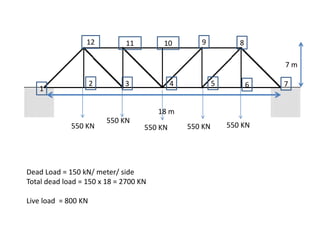
coordinate=[0 0 ;3 0 ;6 0;9 0; 12 0; 15 0; 18 0;15 7; 12 7; 9 7; 6 7; 3 7;];
connectivity=[1 2;2 3;3 4;4 5;5 6; 6 7; 7 8;8 9; 9 10;10 11; 11 12;1
12; 2 12;3 12;3 11; 4 11;4 9; 4 10; 5 9; 5 8;6 8; 7 8];
boundary=[1 1 ; 0 0 ; 0 0 ;0 0 ;0 0;0 0; 1 1;0 0; 0 0;0 0;0 0;0 0];
load=[0 0 0 -550e3 0 -550e3 0 -550e3 0 -550e3 0 -550e3 0 0 0 0 0 0
0 0 0 0 0 0];
Elasticity=2.E+11*[1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1];](https://image.slidesharecdn.com/pratttrussoptimizationusing-161116063529/85/Pratt-truss-optimization-using-14-320.jpg)