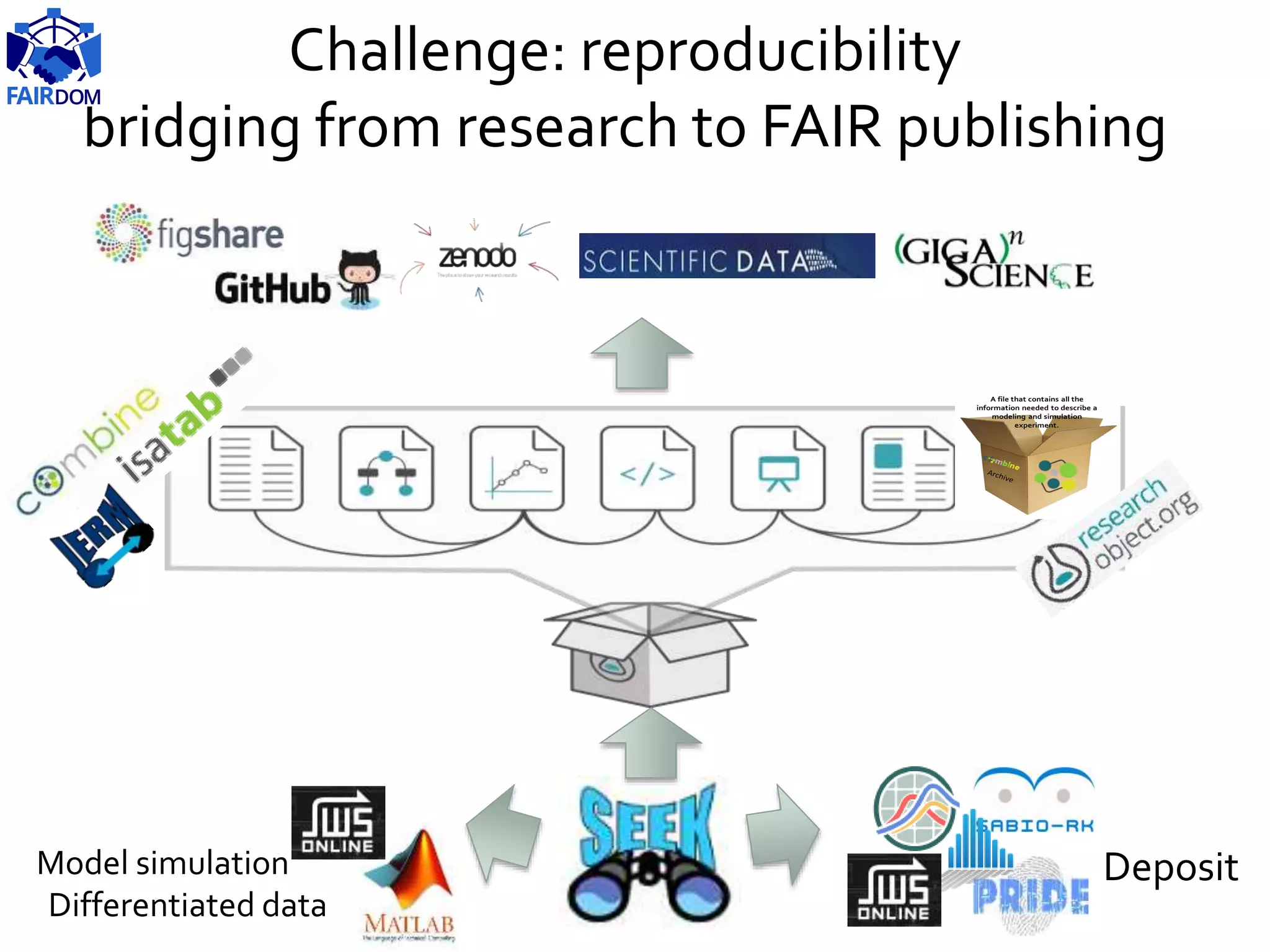
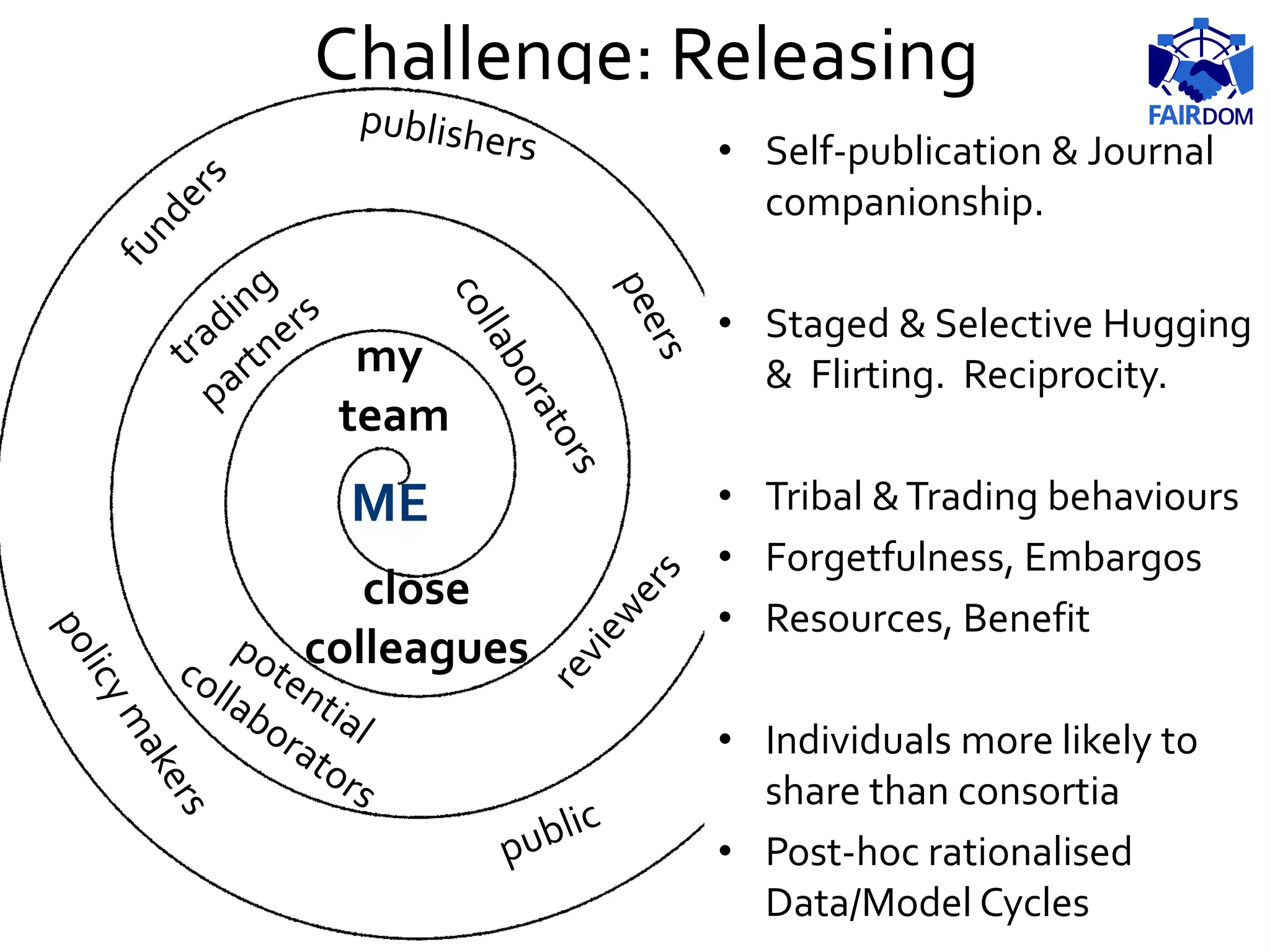
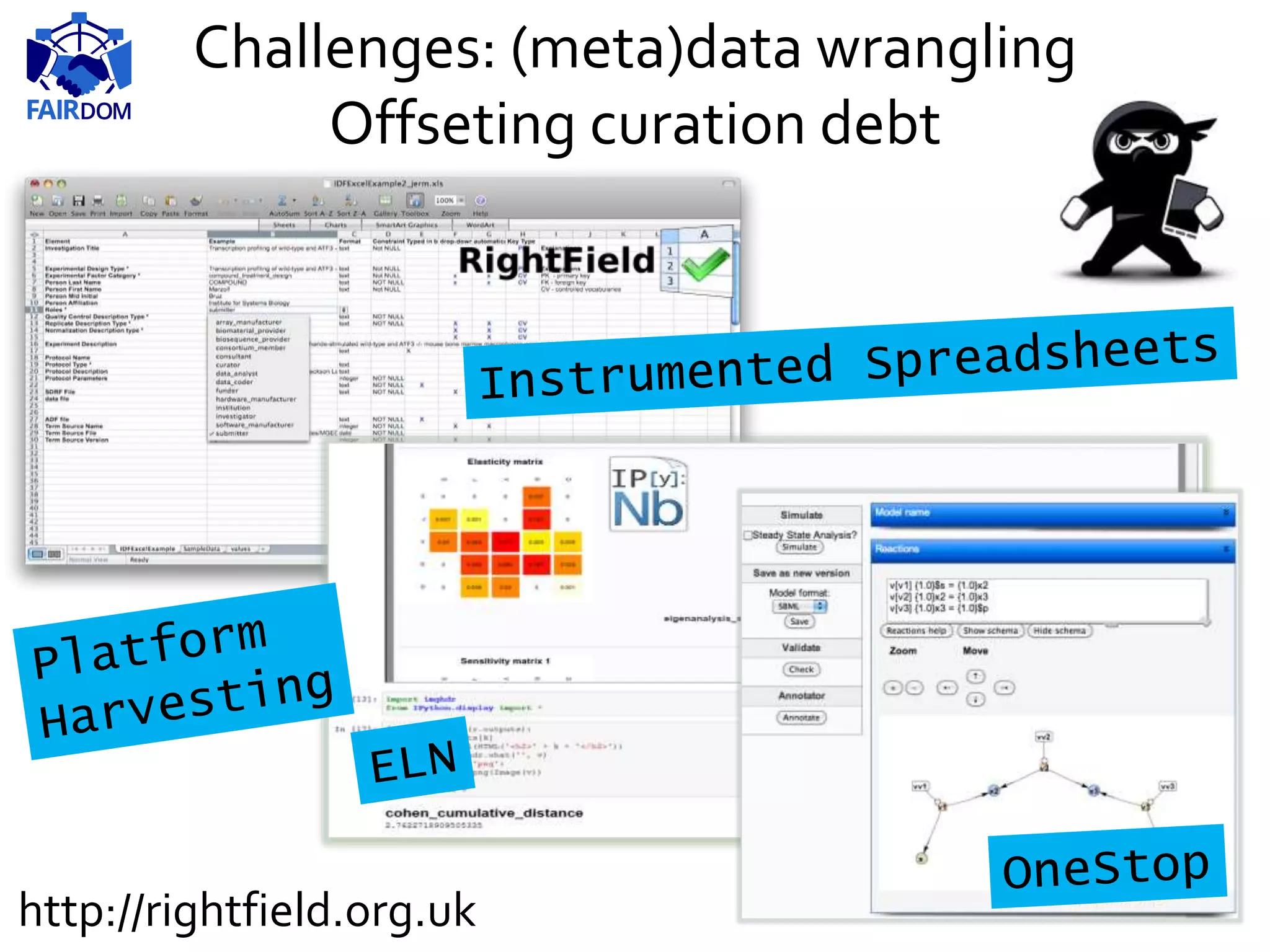
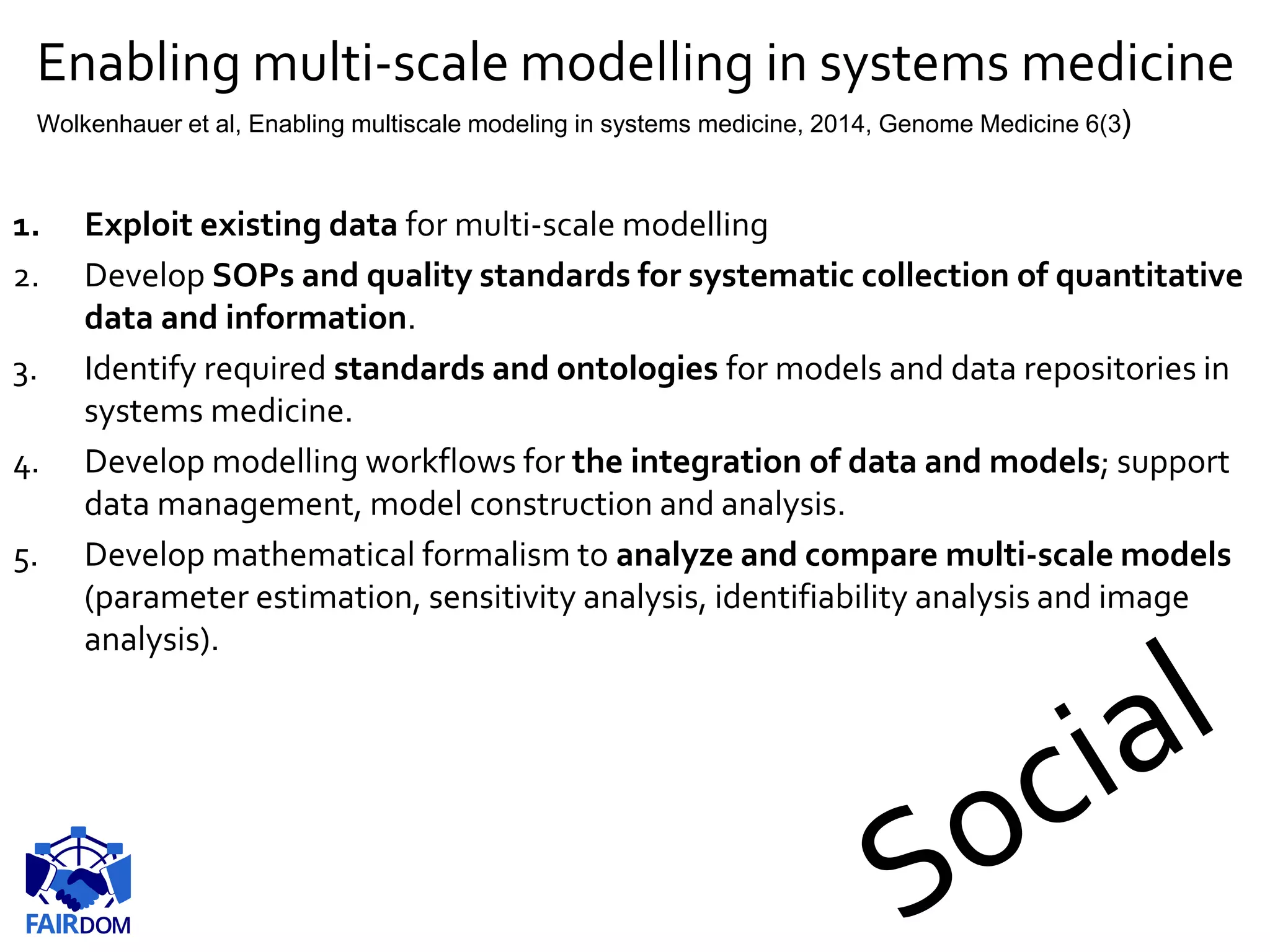
The document highlights the importance of FAIR (Findable, Accessible, Interoperable, Reusable) principles in the management of data and models within systems biology projects. It discusses the challenges and solutions related to reproducibility, data standards, and collaboration, aiming to enhance data sharing and transparency in research. The FAIRDOM initiative and various tools are presented as resources to facilitate effective data management and model sharing among researchers.









![Challenge: Most quantitative databases provide kinetic constants
for enzymes, sometimes binding constants….
Little to help building quantitative descriptions, i.e. concentrations,
sizes, diffusions….
Exceptions: gene expression data, proteomics, metabolomics.
Localisation: The average concentration of a protein in a piece of
brain is of limited use (mix of tissues and subcellular compartments)
[Nicolas Le Novere, 2015]
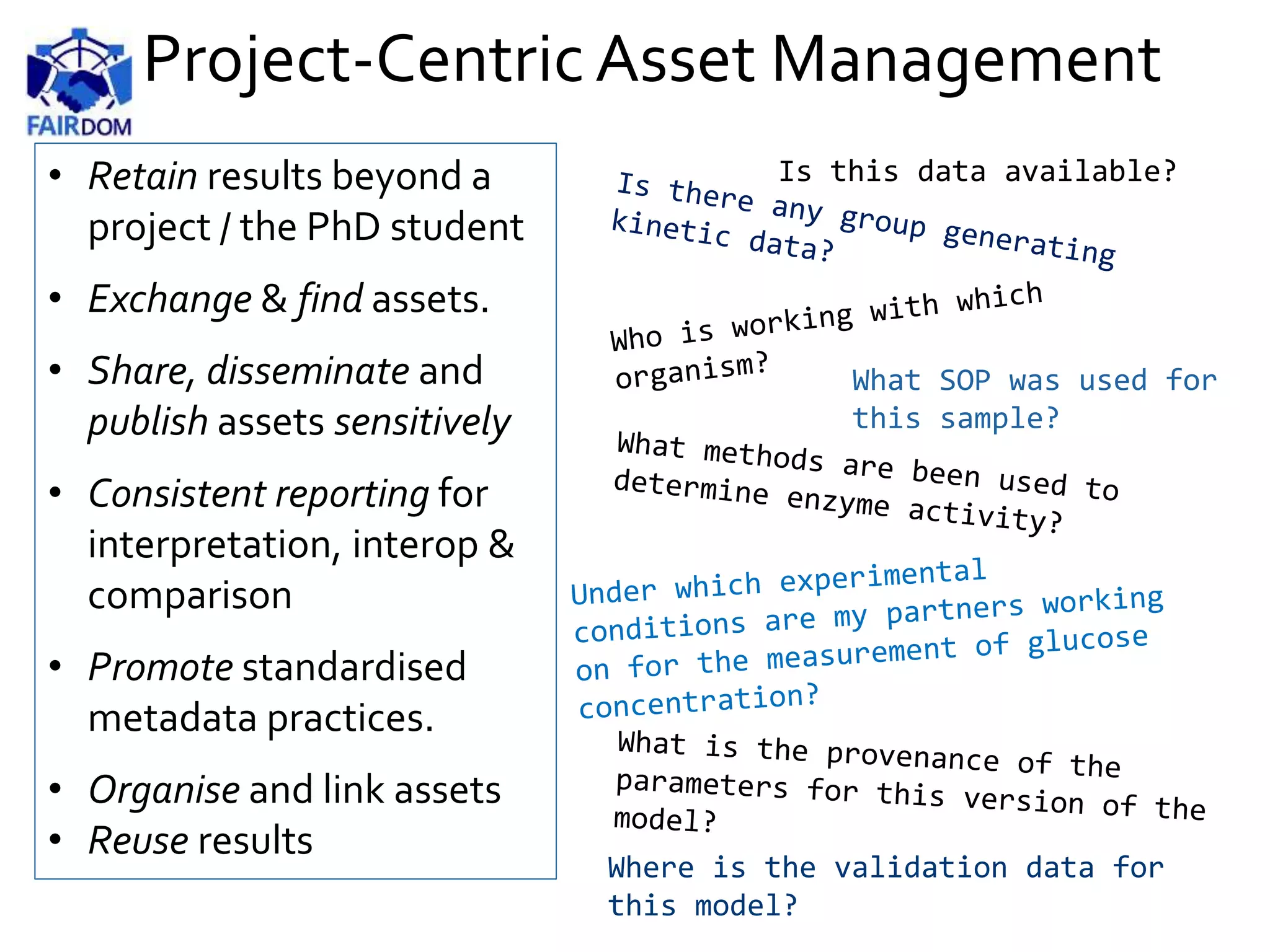
Public-Centric Asset Management
public archives](https://image.slidesharecdn.com/multiscale-biology-2015-final-150721163850-lva1-app6891-151102085423-lva1-app6892/75/FAIR-data-and-model-management-for-systems-biology-and-SOPs-too-12-2048.jpg)




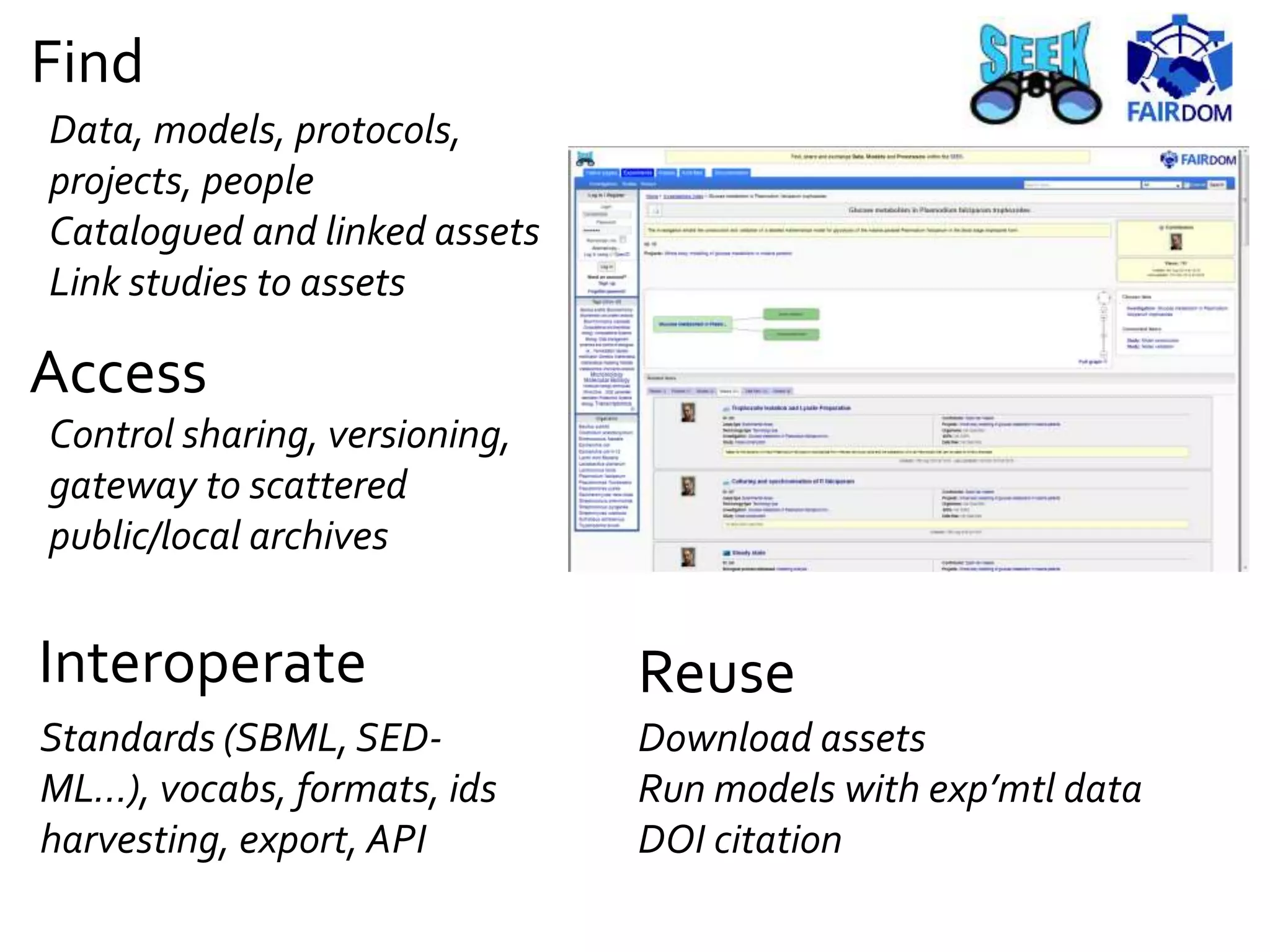
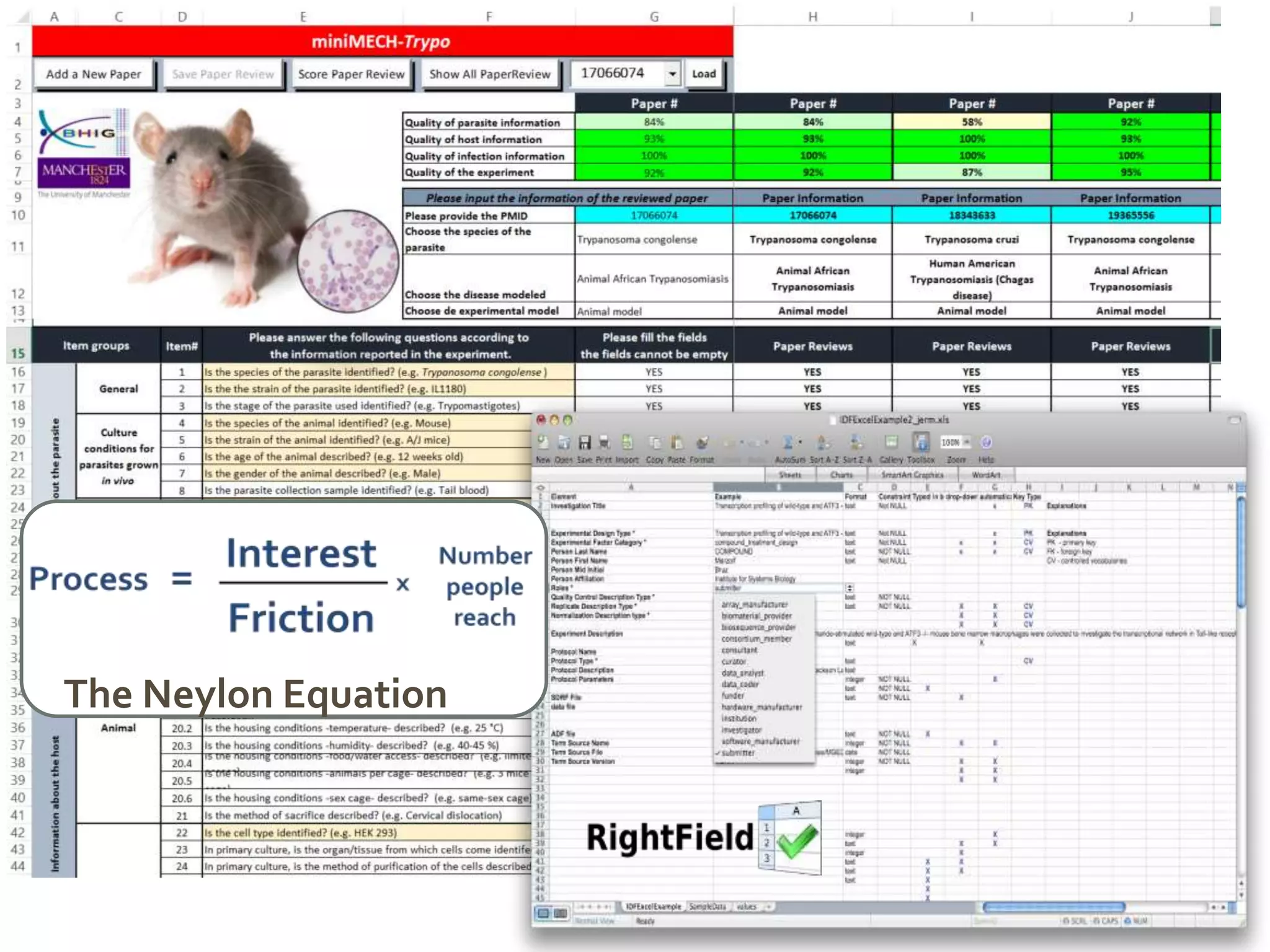





![[Snoep, 2015]
https://doi.org/10.15490/seek.1.investigation.56](https://image.slidesharecdn.com/multiscale-biology-2015-final-150721163850-lva1-app6891-151102085423-lva1-app6892/75/FAIR-data-and-model-management-for-systems-biology-and-SOPs-too-25-2048.jpg)


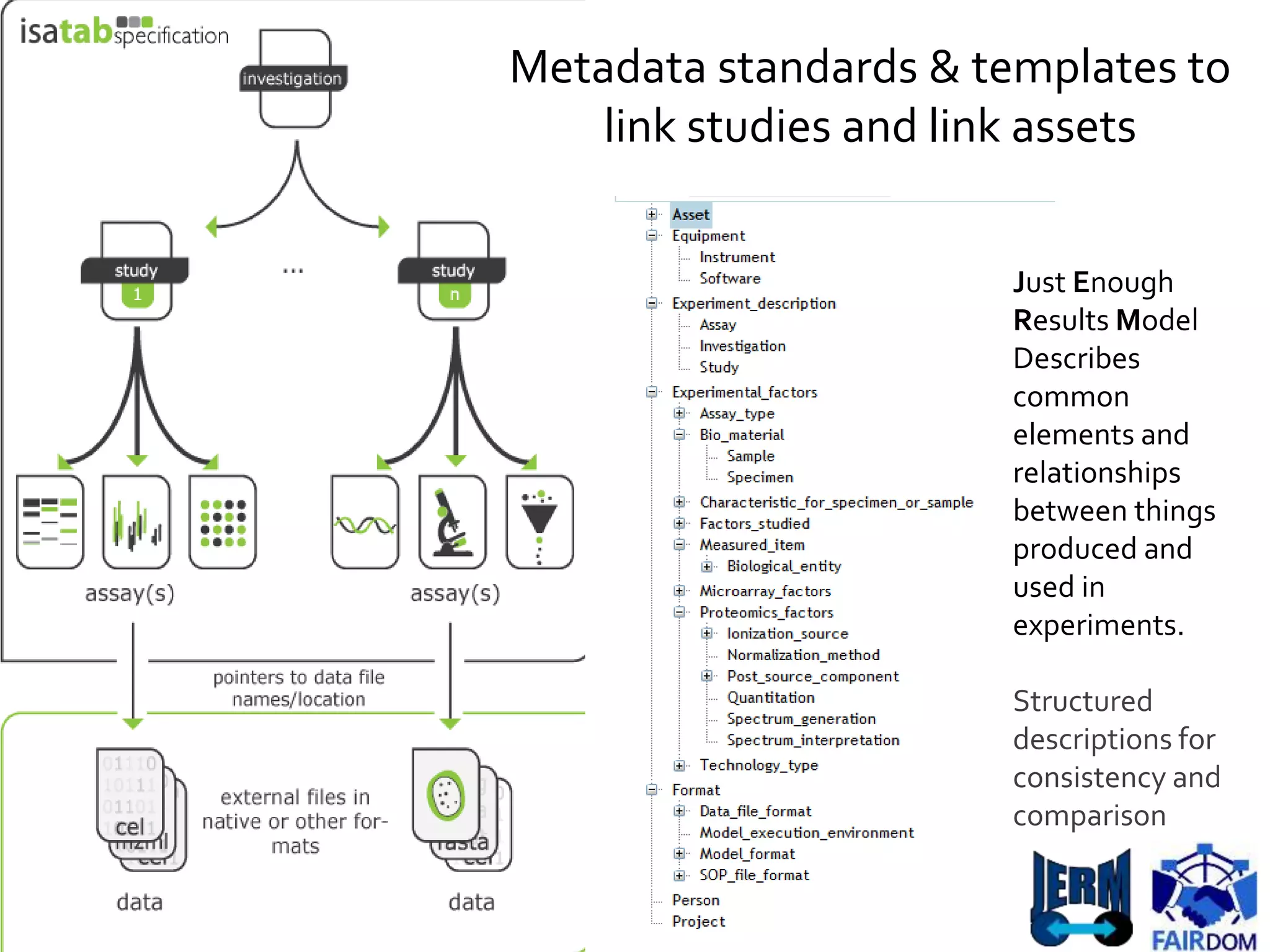
![NuML
[Adapted, Le Novere]](https://image.slidesharecdn.com/multiscale-biology-2015-final-150721163850-lva1-app6891-151102085423-lva1-app6892/75/FAIR-data-and-model-management-for-systems-biology-and-SOPs-too-29-2048.jpg)








![Challenge: model evolution
BiVeS tool: diff in versions of computational models
Provenance,Versioning, Parameter tracking
Releasing updated versions into the literature
Identifying, Interpreting, and CommunicatingChanges in XML-encoded Models of Biological Systems Scharm et. al.
2015, under revision at BIOINFORMATICS
Haus et al, BMC
Systems Biology, 2011, 5:10
Solvent production by
Clostridium acetobutylicum
[Martin Scharm]](https://image.slidesharecdn.com/multiscale-biology-2015-final-150721163850-lva1-app6891-151102085423-lva1-app6892/75/FAIR-data-and-model-management-for-systems-biology-and-SOPs-too-39-2048.jpg)
![F1000Research Living Figures,
versioned articles, in-article data manipulation
R Lawrence Force2015, Vision Award Runner Up http://f1000.com/posters/browse/summary/1097482
Simply data + code
Can change the definition of
a figure, and ultimately the
journal article
Colomb J and Brembs B.
Sub-strains of Drosophila Canton-S differ
markedly in their locomotor behavior [v1;
ref status: indexed, http://f1000r.es/3is]
F1000Research 2014, 3:176
Other labs can replicate the study, or
contribute their data to a meta-analysis
or disease model - figure automatically
updates.
Data updates time-stamped.
New conclusions added via versions.](https://image.slidesharecdn.com/multiscale-biology-2015-final-150721163850-lva1-app6891-151102085423-lva1-app6892/75/FAIR-data-and-model-management-for-systems-biology-and-SOPs-too-40-2048.jpg)