https://imatge.upc.edu/web/publications/active-deep-learning-medical-imaging-segmentation
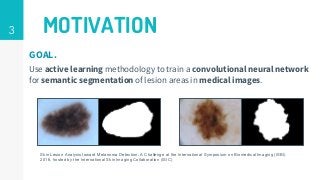
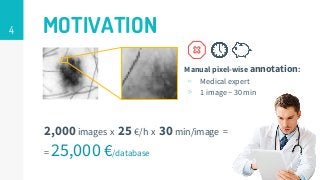
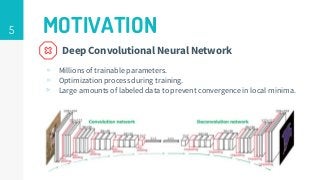
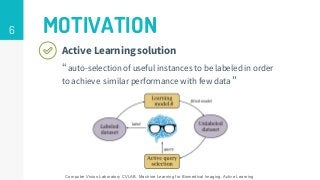
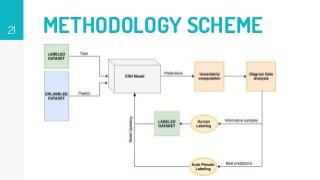
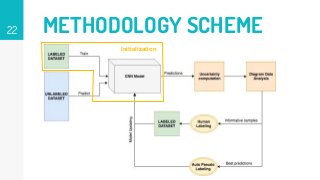
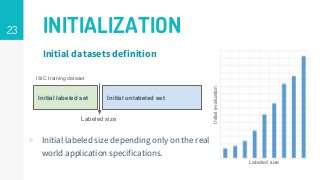
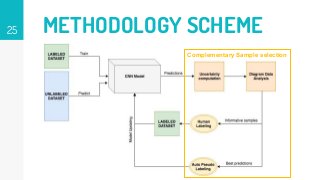
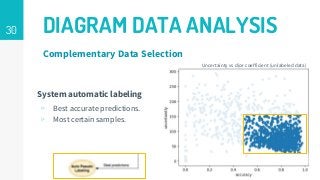
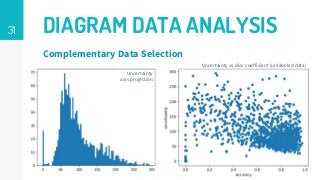
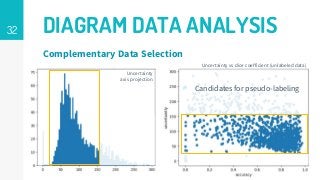
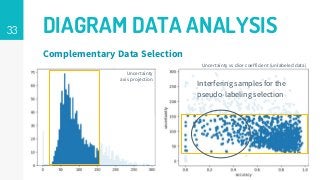
This thesis proposes a novel active learning framework capable to train eectively a convolutional neural network for semantic segmentation of medical imaging, with a limited amount of training labeled data. Our approach tries to apply in segmentation existing active learning techniques, which is becoming an important topic today because of the many problems caused by the lack of large amounts of data. We explore dierent strategies to study the image information and introduce a previously used cost-eective active learning method based on the selection of high condence predictions to assign automatically pseudo-labels with the aim of reducing the manual annotations. First, we made a simple application for handwritten digit classication to get started to the methodology and then we test the system with a medical image database for the treatment of melanoma skin cancer. Finally, we compared the traditional training methods with our active learning proposals, specifying the conditions and parameters required for it to be optimal.