This document discusses selective inference and single-cell differential analysis. It introduces the problem of "double dipping" in the standard single-cell analysis pipeline where the same dataset is used for clustering and differential analysis. Two approaches for addressing this are presented: 1) A method that perturbs clusters before testing for differences, and 2) A test based on a truncated distribution that assumes clusters and genes are given separately. Experiments applying these methods to real single-cell datasets are described. The document outlines challenges in extending these approaches to more complex analyses.

![Standard single-cell analysis pipeline and double dipping
Image taken from [Fang et al., 2021]
Club Single-Cell
February 7th, 2022 / Nathalie Vialaneix
p. 3](https://image.slidesharecdn.com/2022-02-07clubsc-220207161137/85/Selective-inference-and-single-cell-differential-analysis-3-320.jpg)
![Standard single-cell analysis pipeline and double dipping
Image taken from [Fang et al., 2021]
here: differential analysis
Dataset is used twice: (clustering
then differential analysis)
Club Single-Cell
February 7th, 2022 / Nathalie Vialaneix
p. 3](https://image.slidesharecdn.com/2022-02-07clubsc-220207161137/85/Selective-inference-and-single-cell-differential-analysis-4-320.jpg)

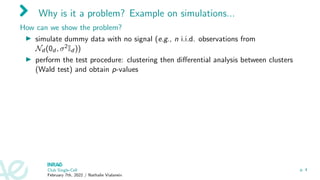
![Why is it a problem? Example on simulations...
How can we show the problem?
I simulate dummy data with no signal (e.g., n i.i.d. observations from
Nd (0d , σ2Id ))
I perform the test procedure: clustering then differential analysis between clusters
(Wald test) and obtain p-values
I What do we expect? Since there is no signal in the data (no true clusters so no
marker genes), p-values ∼ U[0, 1]
Club Single-Cell
February 7th, 2022 / Nathalie Vialaneix
p. 4](https://image.slidesharecdn.com/2022-02-07clubsc-220207161137/85/Selective-inference-and-single-cell-differential-analysis-7-320.jpg)
![First question [Gao et al., 2021]
Is the average value of vector X in first cluster different of what it is in the section
cluster?
Club Single-Cell
February 7th, 2022 / Nathalie Vialaneix
p. 5](https://image.slidesharecdn.com/2022-02-07clubsc-220207161137/85/Selective-inference-and-single-cell-differential-analysis-8-320.jpg)
![First question using a train/test approach [Gao et al., 2021]
Is the average value of vector X, in first cluster different of what it is in the second
cluster?
Club Single-Cell
February 7th, 2022 / Nathalie Vialaneix
p. 6](https://image.slidesharecdn.com/2022-02-07clubsc-220207161137/85/Selective-inference-and-single-cell-differential-analysis-9-320.jpg)
![Second question (at the level of marker gene)
[Zhang et al., 2019]
Is the average expression of a given gene, xj , in first cluster different of what it is in
the second cluster?
Club Single-Cell
February 7th, 2022 / Nathalie Vialaneix
p. 7](https://image.slidesharecdn.com/2022-02-07clubsc-220207161137/85/Selective-inference-and-single-cell-differential-analysis-10-320.jpg)


![Question 1 [Gao et al., 2021]
Denoting by D := kX(1) − X(2)k and φ a rv from χ2 (with parameters depending on
X), define a perturbed version of the data that:
I pulls clusters apart if φ > D
I push clusters together if φ < D
There is a way to obtain a valid p-value from the distribution of obtained clusters (that
depends on the rv φ).
Club Single-Cell
February 7th, 2022 / Nathalie Vialaneix
p. 10](https://image.slidesharecdn.com/2022-02-07clubsc-220207161137/85/Selective-inference-and-single-cell-differential-analysis-13-320.jpg)
![Question 1 [Gao et al., 2021]
Is it usable? More or less...
1. either: you have a way to have a explicit description of the perturbed cluster
definition
Club Single-Cell
February 7th, 2022 / Nathalie Vialaneix
p. 11](https://image.slidesharecdn.com/2022-02-07clubsc-220207161137/85/Selective-inference-and-single-cell-differential-analysis-14-320.jpg)
![Question 1 [Gao et al., 2021]
Is it usable? More or less...
1. either: you have a way to have a explicit description of the perturbed cluster
definition
Only available for HC in [Gao et al., 2021].
Club Single-Cell
February 7th, 2022 / Nathalie Vialaneix
p. 11](https://image.slidesharecdn.com/2022-02-07clubsc-220207161137/85/Selective-inference-and-single-cell-differential-analysis-15-320.jpg)
![Question 1 [Gao et al., 2021]
Is it usable? More or less...
1. either: you have a way to have a explicit description of the perturbed cluster
definition
Only available for HC in [Gao et al., 2021].
2. or: you simulate the distribution (using random draws of φ)
Club Single-Cell
February 7th, 2022 / Nathalie Vialaneix
p. 11](https://image.slidesharecdn.com/2022-02-07clubsc-220207161137/85/Selective-inference-and-single-cell-differential-analysis-16-320.jpg)
![Question 1 [Gao et al., 2021]
Is it usable? More or less...
1. either: you have a way to have a explicit description of the perturbed cluster
definition
Only available for HC in [Gao et al., 2021].
2. or: you simulate the distribution (using random draws of φ)
But you need to have plenty of time.
Club Single-Cell
February 7th, 2022 / Nathalie Vialaneix
p. 11](https://image.slidesharecdn.com/2022-02-07clubsc-220207161137/85/Selective-inference-and-single-cell-differential-analysis-17-320.jpg)
![Question 1 [Gao et al., 2021]
Is it usable? More or less...
1. either: you have a way to have a explicit description of the perturbed cluster
definition
Only available for HC in [Gao et al., 2021].
2. or: you simulate the distribution (using random draws of φ)
But you need to have plenty of time.
The method is available as an R package: clusterpval
https://www.lucylgao.com/clusterpval/
Club Single-Cell
February 7th, 2022 / Nathalie Vialaneix
p. 11](https://image.slidesharecdn.com/2022-02-07clubsc-220207161137/85/Selective-inference-and-single-cell-differential-analysis-18-320.jpg)
![Experiment
Data from [Zheng et al., 2017] with clustering of peripheral blood mononuclear cells
prior to sequencing (antibody-based bead enrichment + fluorescent activated cell
sorting) ⇒ ground truth
Club Single-Cell
February 7th, 2022 / Nathalie Vialaneix
p. 12](https://image.slidesharecdn.com/2022-02-07clubsc-220207161137/85/Selective-inference-and-single-cell-differential-analysis-19-320.jpg)
![Experiment
Data from [Zheng et al., 2017] with clustering of peripheral blood mononuclear cells
prior to sequencing (antibody-based bead enrichment + fluorescent activated cell
sorting) ⇒ ground truth
Derivation of:
I negative control (selection of 600 memory T cells)
I positive control (selection of 200 memory T cells + 200 B cells + monocytes)
Club Single-Cell
February 7th, 2022 / Nathalie Vialaneix
p. 12](https://image.slidesharecdn.com/2022-02-07clubsc-220207161137/85/Selective-inference-and-single-cell-differential-analysis-20-320.jpg)
![Experiment
Data from [Zheng et al., 2017] with clustering of peripheral blood mononuclear cells
prior to sequencing (antibody-based bead enrichment + fluorescent activated cell
sorting) ⇒ ground truth
Derivation of:
I negative control (selection of 600 memory T cells)
I positive control (selection of 200 memory T cells + 200 B cells + monocytes)
Method: clustering with HAC (3 clusters) then differential analysis (Wald test versus
their test)
Club Single-Cell
February 7th, 2022 / Nathalie Vialaneix
p. 12](https://image.slidesharecdn.com/2022-02-07clubsc-220207161137/85/Selective-inference-and-single-cell-differential-analysis-21-320.jpg)


![Question 2 [Zhang et al., 2019]
Use a test based on a truncated distribution
Club Single-Cell
February 7th, 2022 / Nathalie Vialaneix
p. 15](https://image.slidesharecdn.com/2022-02-07clubsc-220207161137/85/Selective-inference-and-single-cell-differential-analysis-24-320.jpg)
![Question 2 [Zhang et al., 2019]
Club Single-Cell
February 7th, 2022 / Nathalie Vialaneix
p. 16](https://image.slidesharecdn.com/2022-02-07clubsc-220207161137/85/Selective-inference-and-single-cell-differential-analysis-25-320.jpg)
![Question 2 [Zhang et al., 2019]
Remarks on this approach:
I the separating hyperplane is supposed to be given ⇒ contrains the clustering
method and requires that it is performed on a separate dataset
I genes are supposed to be not correlated (very, very strong assumption...)
I method available as a python tool at
https://github.com/jessemzhang/tn_test
Club Single-Cell
February 7th, 2022 / Nathalie Vialaneix
p. 17](https://image.slidesharecdn.com/2022-02-07clubsc-220207161137/85/Selective-inference-and-single-cell-differential-analysis-26-320.jpg)
![Experiment 1
Again... data from [Zheng et al., 2017]...
Method:
I use SEURAT for clustering (9 clusters)
I use SEURAT and TN for differential analysis between the first two clusters
Club Single-Cell
February 7th, 2022 / Nathalie Vialaneix
p. 18](https://image.slidesharecdn.com/2022-02-07clubsc-220207161137/85/Selective-inference-and-single-cell-differential-analysis-27-320.jpg)
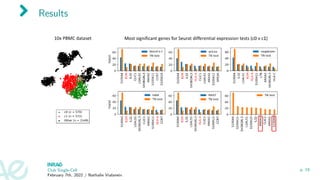
![Experiment 2
Data from [Kolodziejczyk et al., 2015]
Impact of overclustering on results
Club Single-Cell
February 7th, 2022 / Nathalie Vialaneix
p. 20](https://image.slidesharecdn.com/2022-02-07clubsc-220207161137/85/Selective-inference-and-single-cell-differential-analysis-29-320.jpg)
