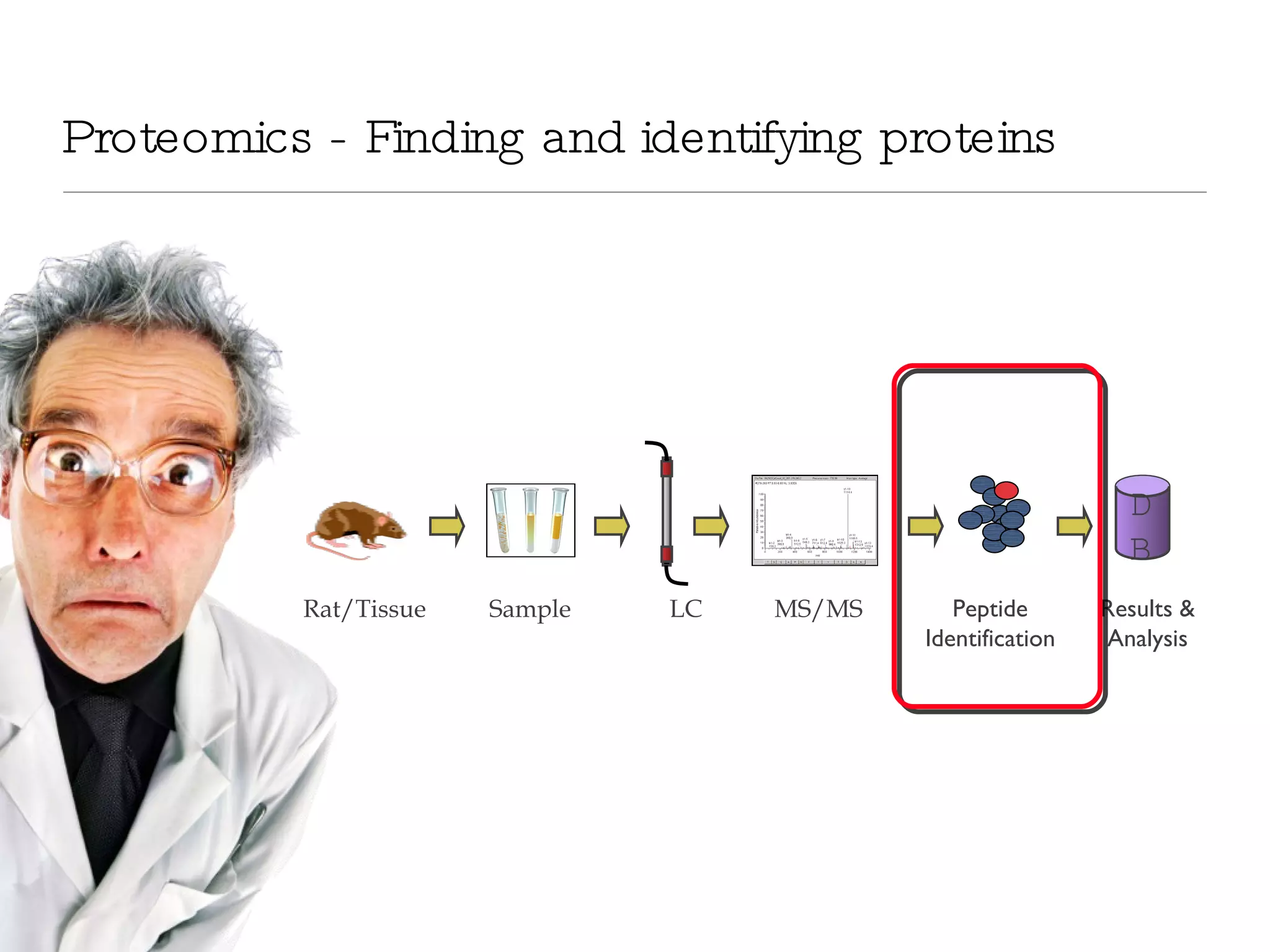
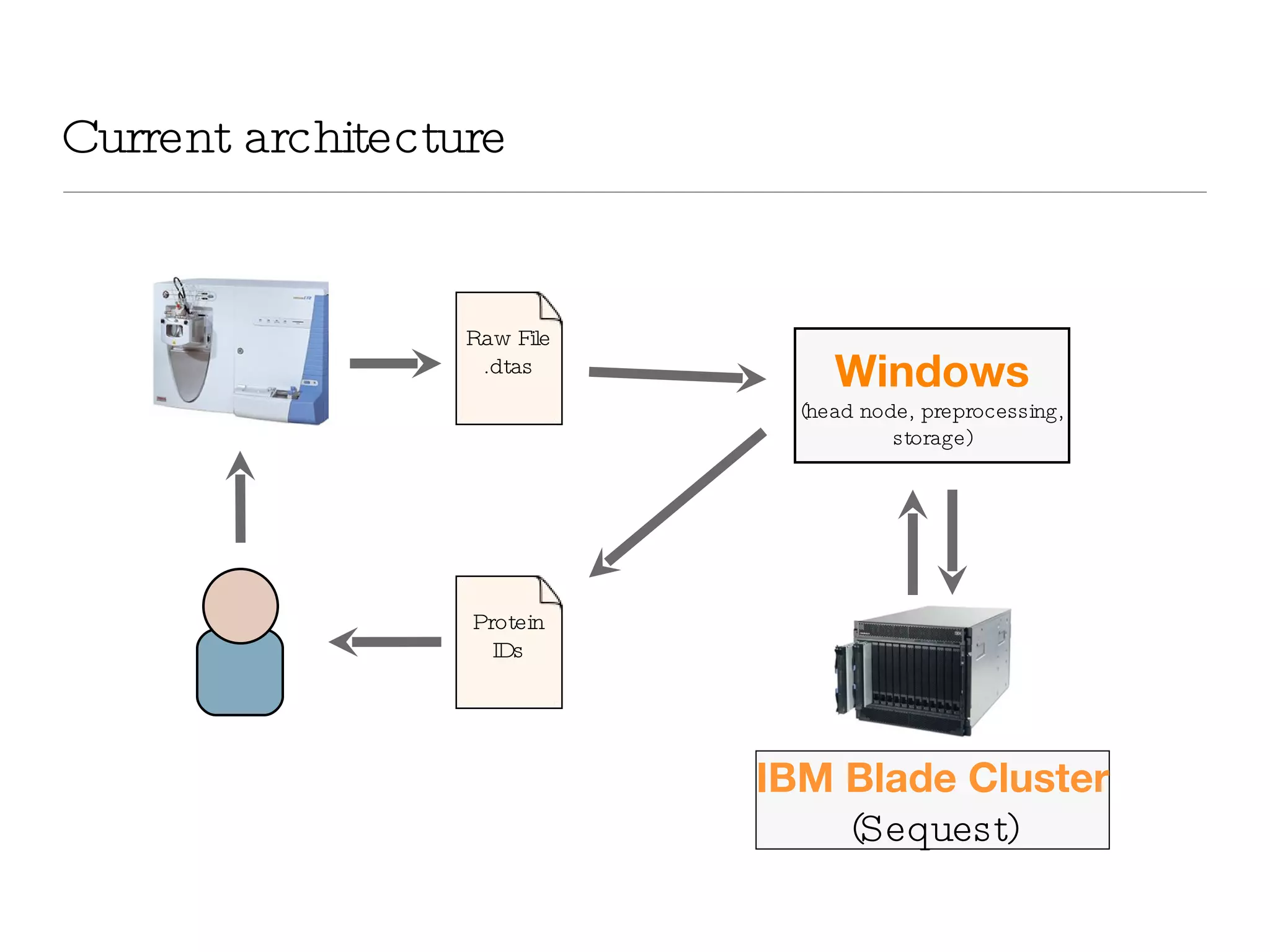
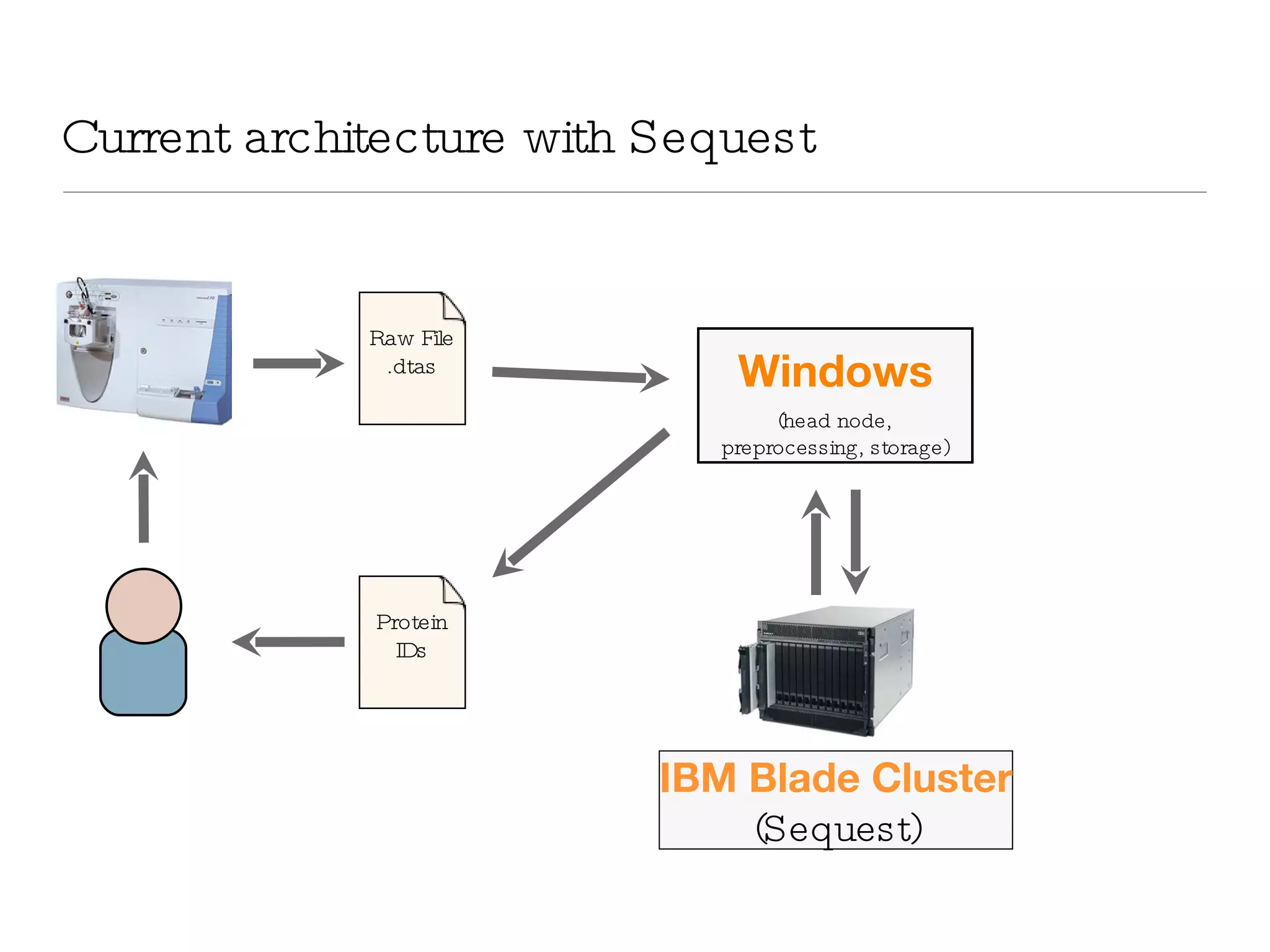
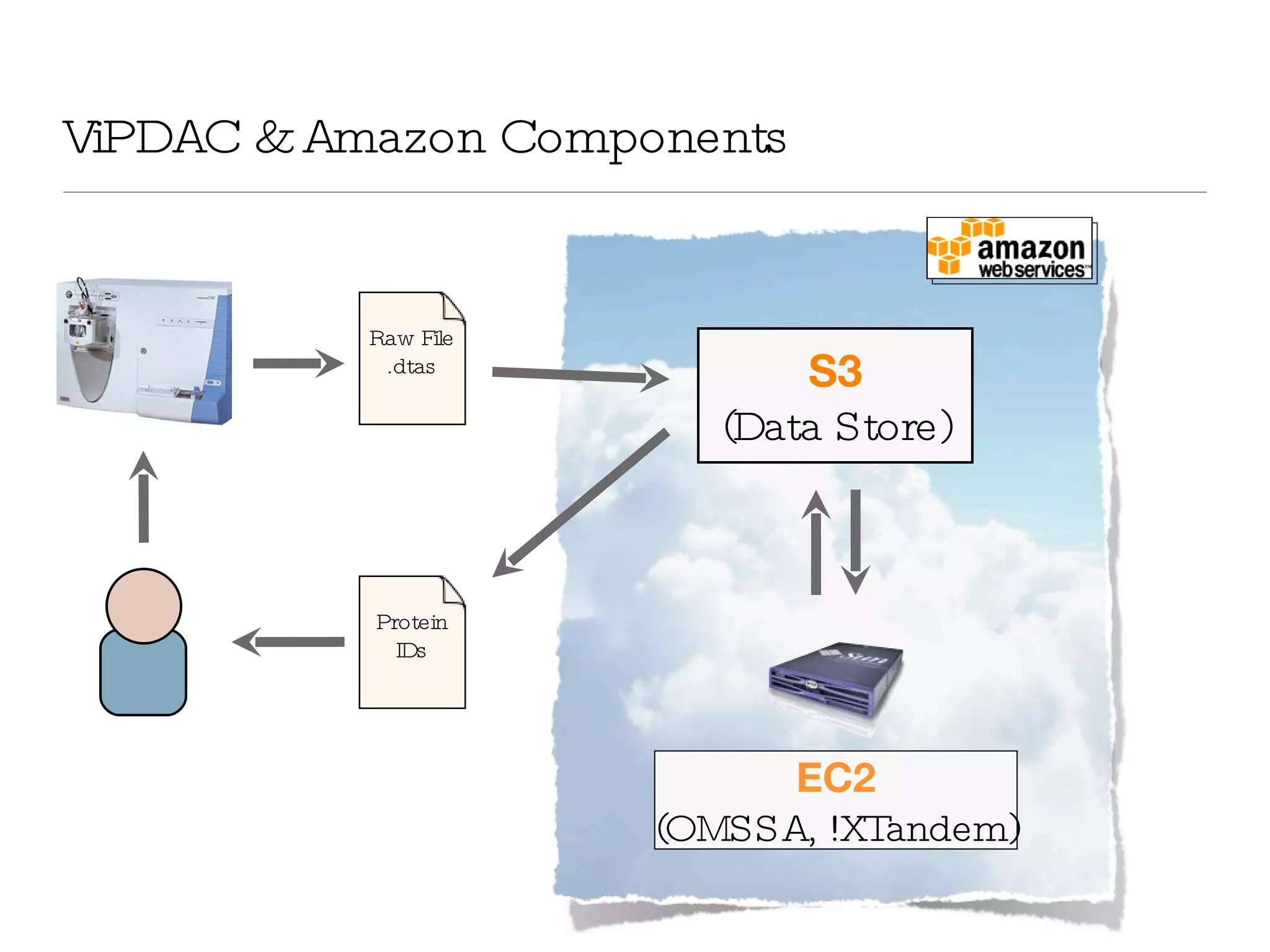
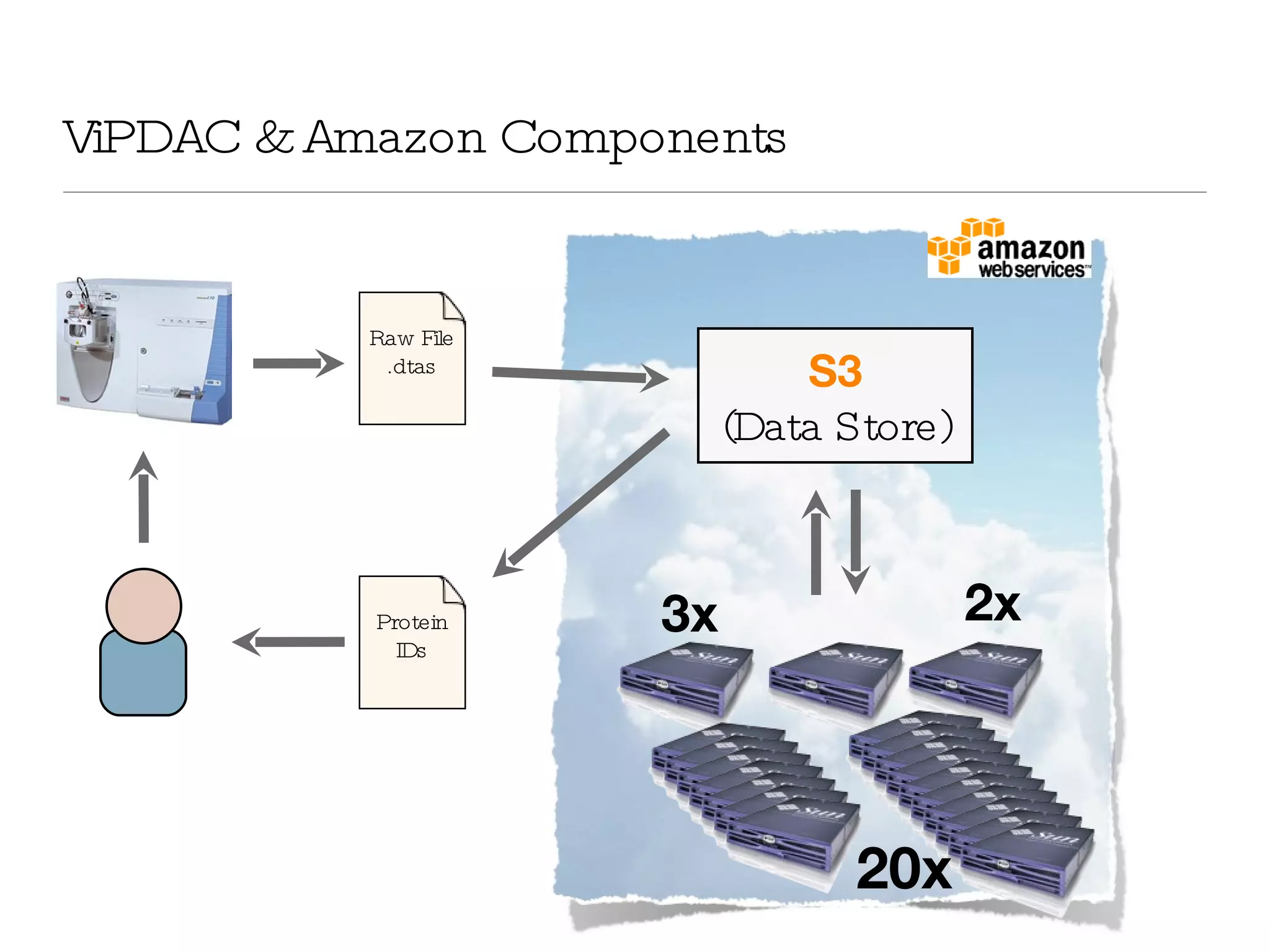
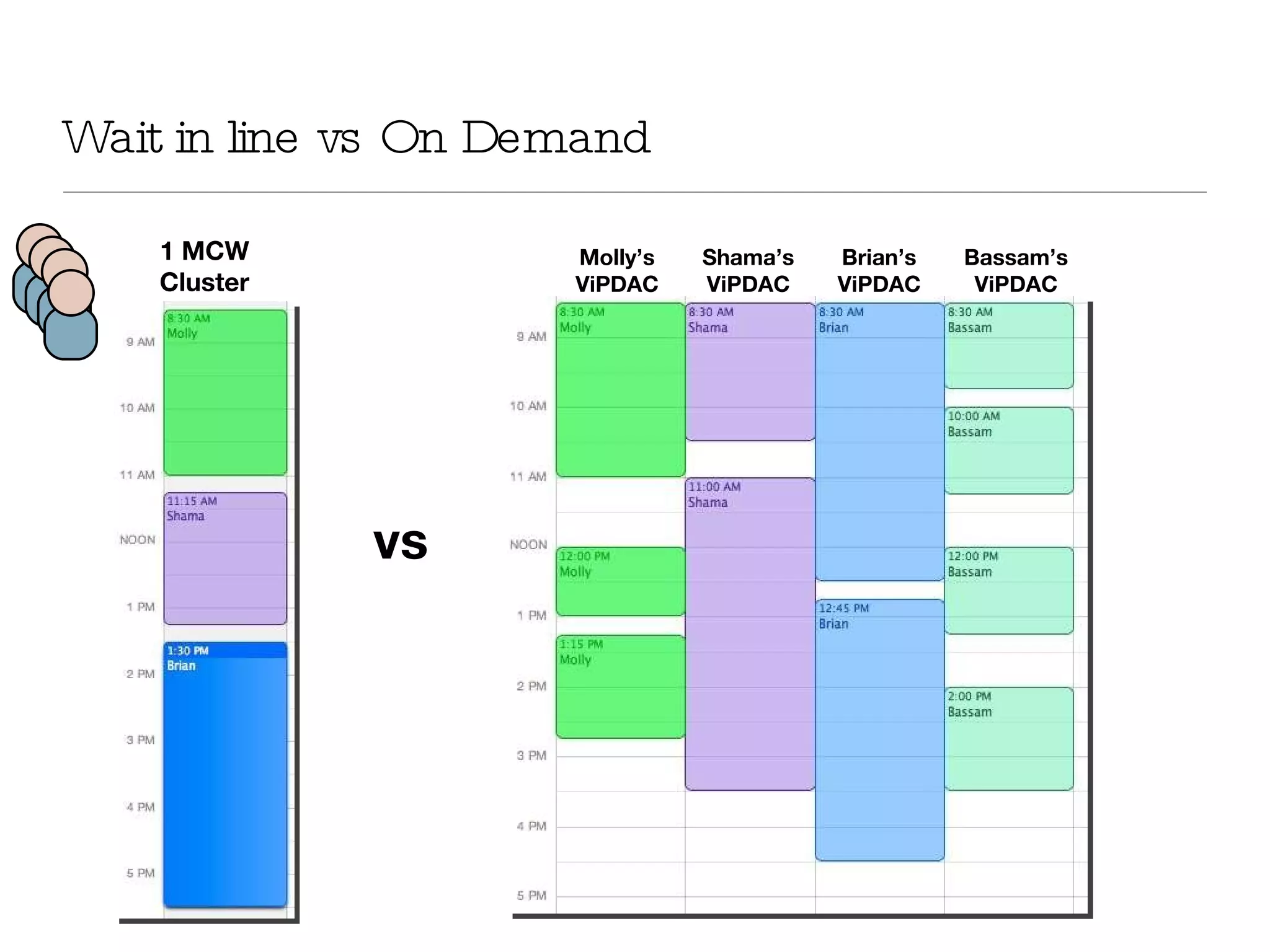
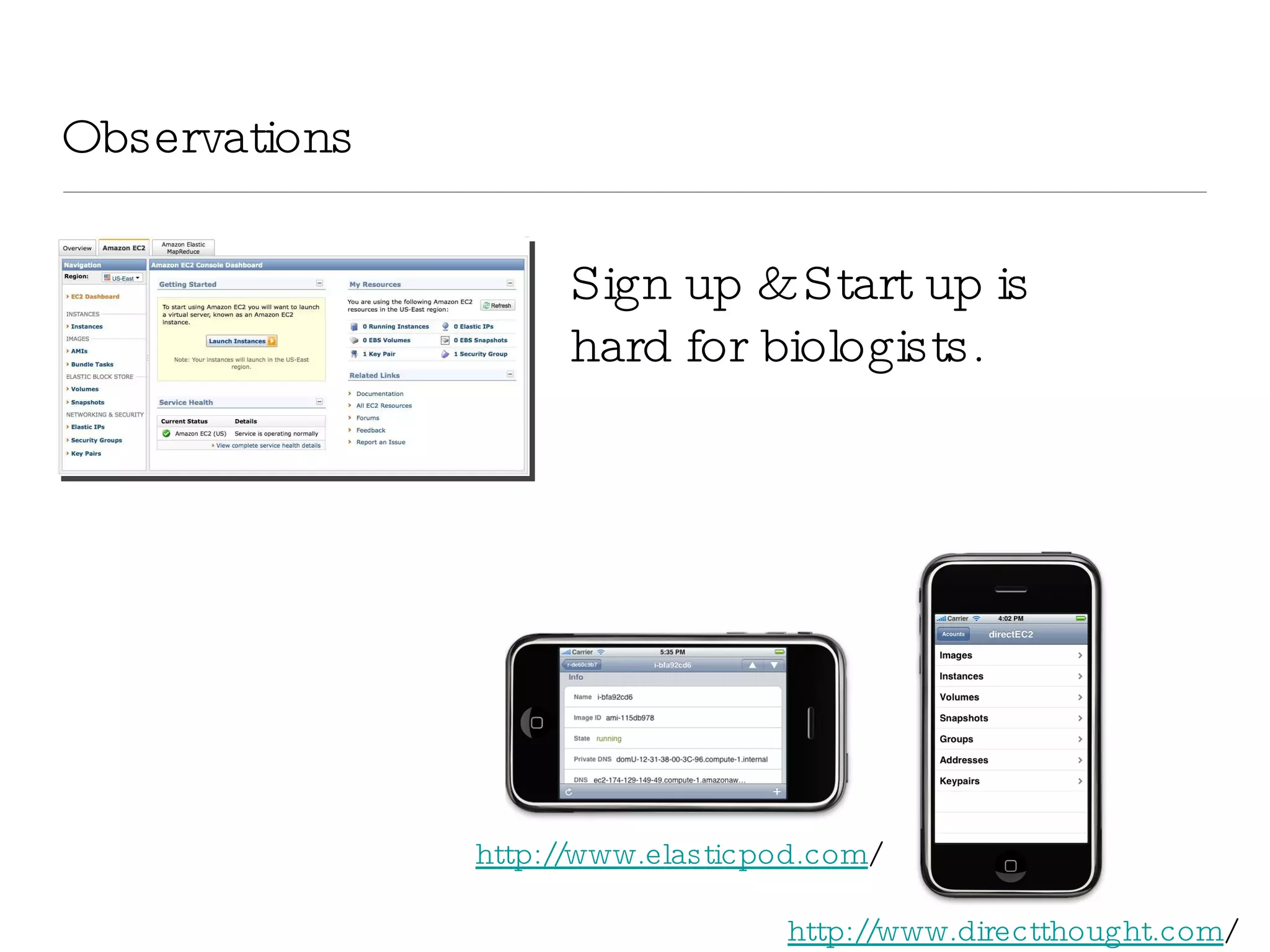
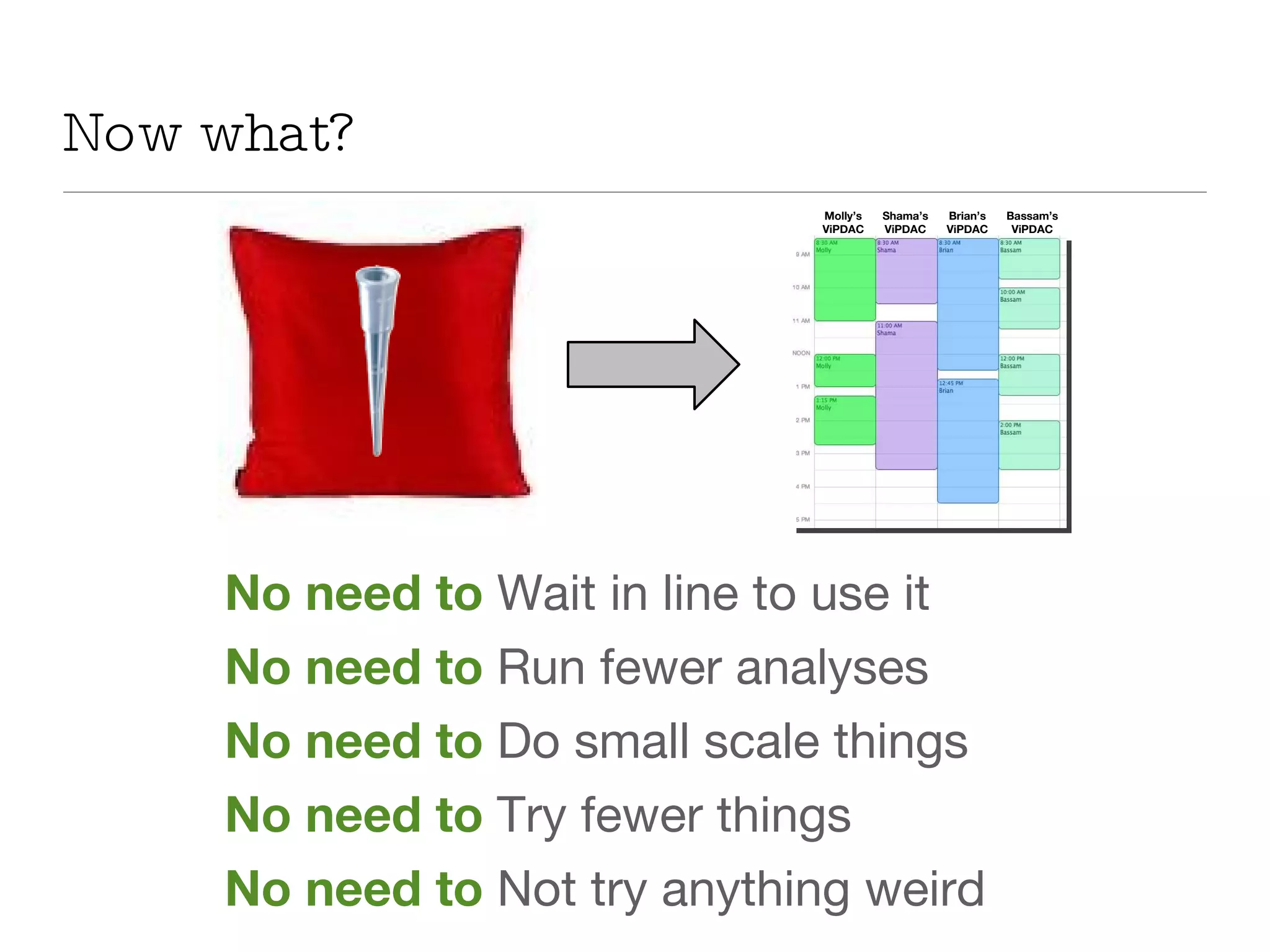
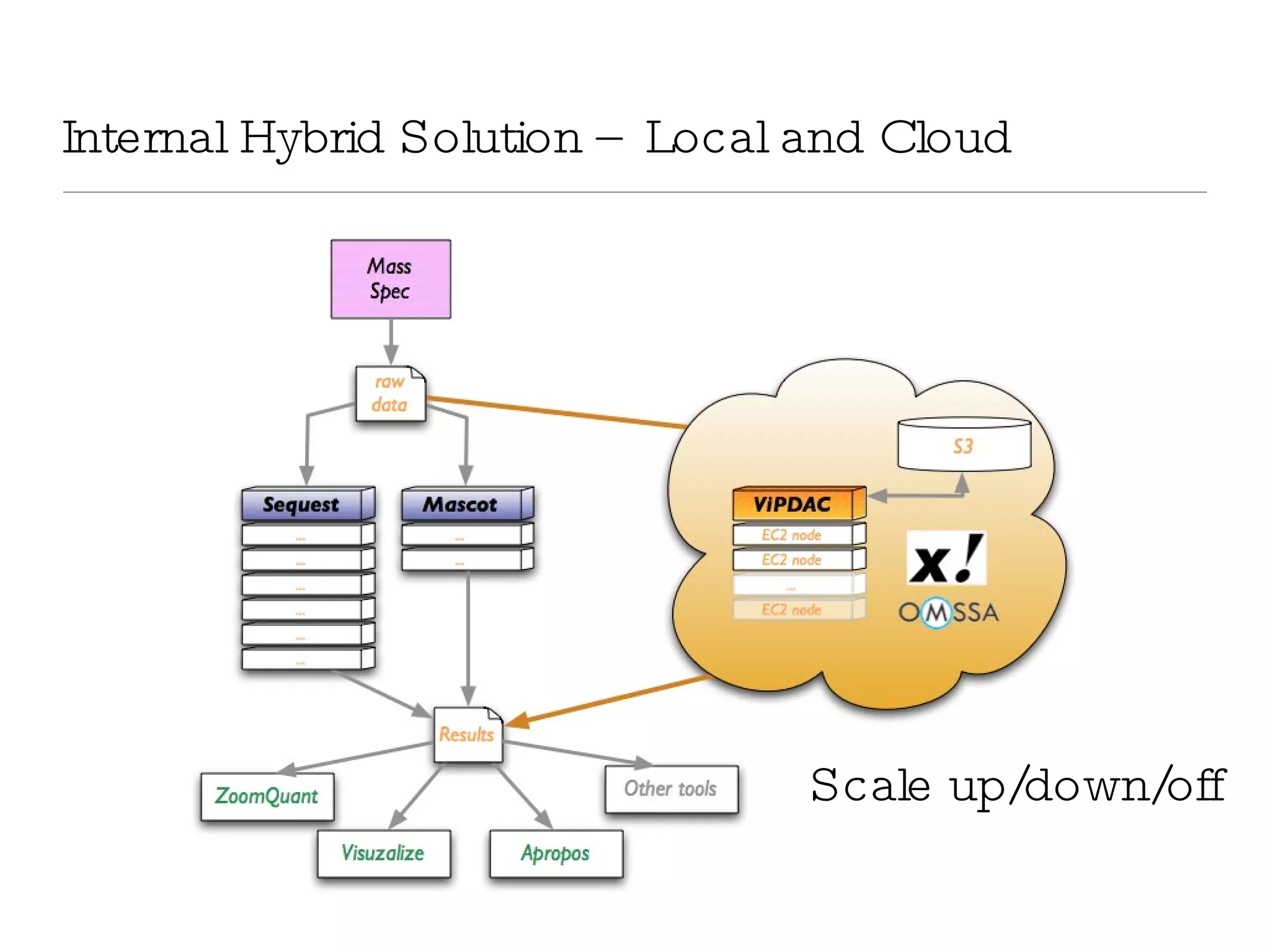
The document introduces VIPDAC, a cloud-based proteomics analysis suite leveraging Amazon Web Services to enhance computational capabilities for researchers. It discusses the limitations of traditional computing resources and the benefits of cloud solutions, such as eliminating wait times and enabling larger-scale analyses. The authors emphasize the accessibility of computational resources for biologists and propose a hybrid model that integrates local and cloud computing.