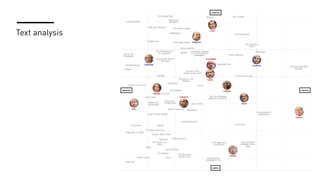
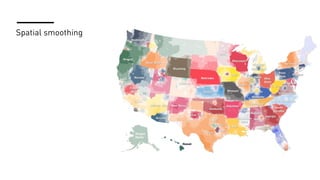
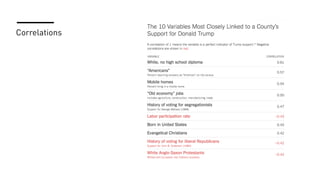
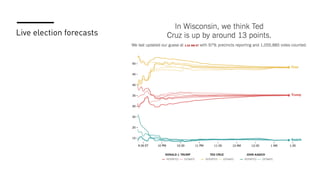
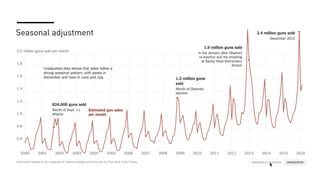
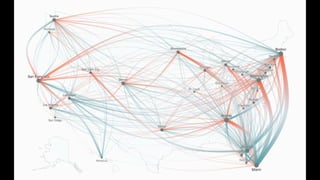
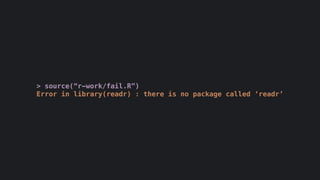
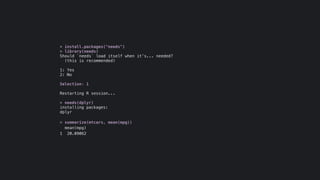
The document discusses how R is used at The New York Times Graphics desk for various tasks like text analysis, spatial smoothing, correlations, live election forecasts, and seasonal adjustment. It provides examples of code for passing data between R and JavaScript, running R scripts asynchronously from JavaScript, and using Makefiles with R. It also notes some potential issues like packages not being installed that could cause errors when running R code.




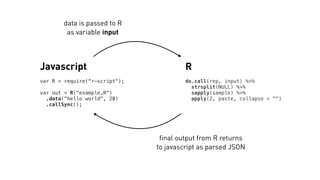
![R
attach(input[[1]])
df %>%
mutate(group = cut(rating, nGroups)) %>%
group_by(group) %>%
summarize_each(funs_(fxn)) %>%
transmute(group = as.character(group),
rating, advance)
Javascript
var R = require(“r-script");
R("example/ex-async.R")
.data({df: attitude,
nGroups: 3,
fxn: "mean" })
.call(function(err, d) {
if (err) throw err;
console.log(d);
});](https://image.slidesharecdn.com/talk-2016-04-09-160511214259/85/Using-R-at-NYT-Graphics-12-320.jpg)







![> library(dplyr)
Attaching package: ‘dplyr’
The Following objects are masked from ‘package:stats’:
filter, lag
The following objects are masked from ‘package:base’:
intersect, setdiff, setequal, union
> library(plyr)
-----------—--------—-------------—--------—-------------—--------—--
You have loaded plyr after dplyr - this is likely to cause problems.
If you need functions from both plyr and dplyr, please load plyr first,
then dplyr:
library(plyr); library(dplyr)
-----------—--------—-------------—--------—-------------—--------—--
Attaching package: ‘plyr’
The following objects are masked from ‘package:dplyr’:
arrange, count, desc, failwith, id, mutate, rename,
summarise, summarize
>iris %>%
+ group_by(Species) %>%
+ summarize(mean(Sepal.Length))
mean(Sepal.Length)
1 5.84333
>
> needs(dplyr, plyr)
> prioritize(dplyr)
> iris %>%
+ group_by(Species) %>%
+ summarize(mean(Sepal.Length))
Source: local data frame [3 x 2]
Species mean(Sepal.Length)
(fctr) (dbl)
1 setosa 5.006
2 versicolor 5.936
3 virginica 6.588
>](https://image.slidesharecdn.com/talk-2016-04-09-160511214259/85/Using-R-at-NYT-Graphics-26-320.jpg)
