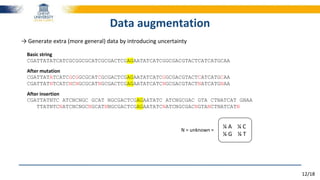
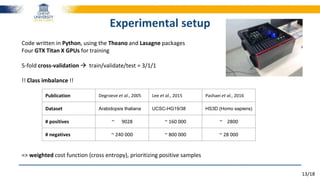
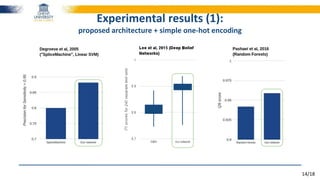
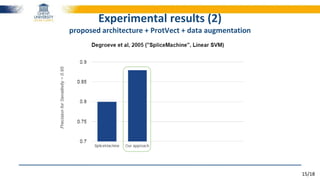
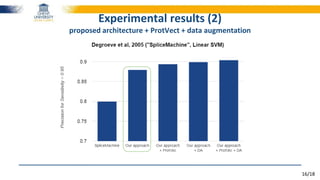
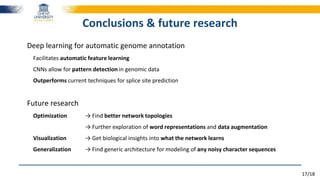
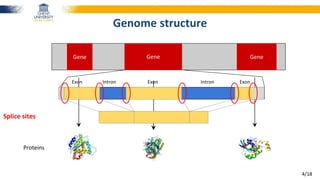
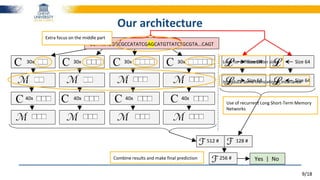
The document discusses a deep learning approach to automatic genome annotation, focusing on identifying gene structures and functionalities using convolutional neural networks (CNNs) and word representations. It highlights the success of this method in surpassing traditional manual techniques, emphasizing data augmentation and the use of the protvec representation for enhancing performance. Future research aims to optimize network architectures and gain biological insights from the findings.

![10/18
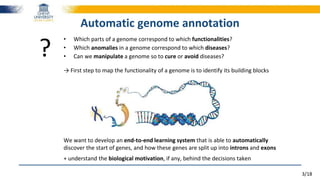
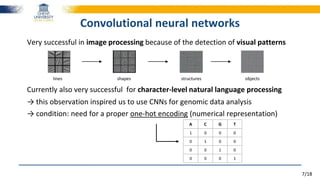
Very important in Natural Language Processing
Each word is represented by a vector of floating-point values
wi = [0.25, 0.30, -0.18, 0.45, …]
Each word vector is trained by its neighbours in a vast collection of texts→ trained to predict its neighbours
The quick brown fox jumps over the lazy dog
Result = powerful word embeddings
Word2Vec representation
King - Man + Woman ≈ Queen
Man
King
Woman
Queen](https://image.slidesharecdn.com/2016-180724144110/85/Towards-reading-genomic-data-using-deep-learning-driven-NLP-techniques-10-320.jpg)
![11/18
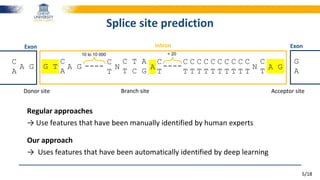
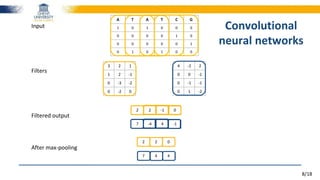
Word2Vec algorithm applied on protein/DNA sequences
Each 𝒏-gram represented by a vector
e.g., GTC = [0.359, 0.211, -0.492, …, 0.129]
Each word vector is trained by its neighbours on a genome → trained to predict its neighbours
... ATG TGT GTC TCA CAC …
ProtVec representation](https://image.slidesharecdn.com/2016-180724144110/85/Towards-reading-genomic-data-using-deep-learning-driven-NLP-techniques-11-320.jpg)