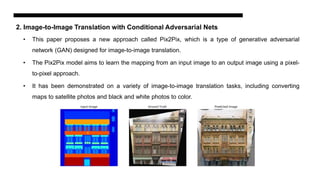
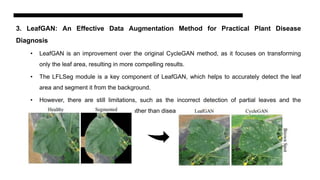
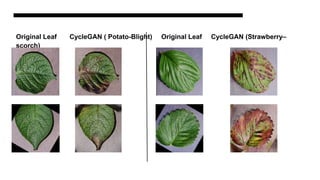
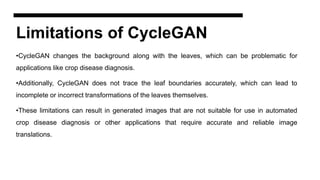
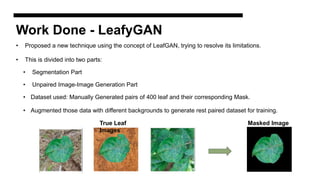
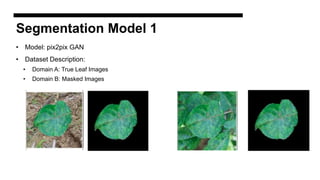
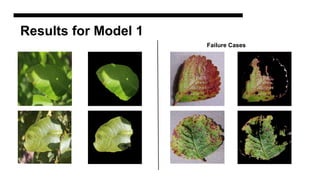
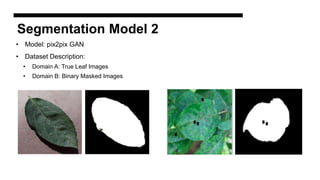
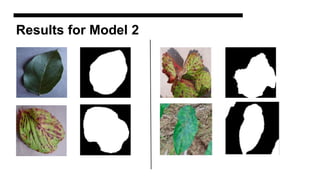
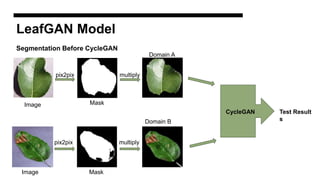
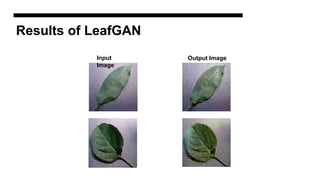
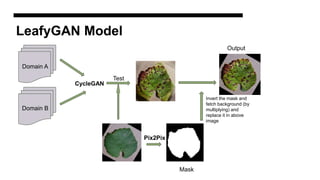
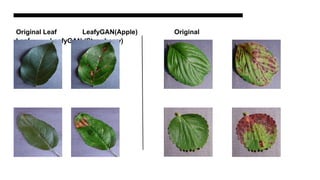
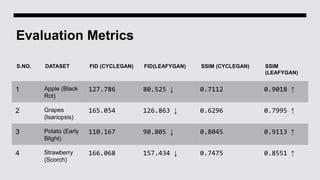
This document discusses data augmentation techniques for improving plant disease diagnosis using deep learning models. It presents work done using CycleGAN and a proposed LeafyGAN model. CycleGAN was able to generate new images but had limitations like changing backgrounds. LeafyGAN segmented leaves first before generation and produced higher quality images with more similar features compared to the originals, as measured by FID and SSIM metrics. The document concludes LeafyGAN may be a promising alternative to CycleGAN for image generation tasks.