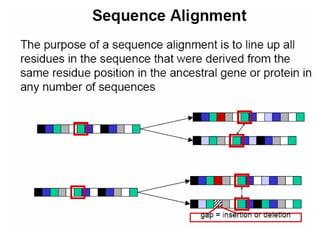
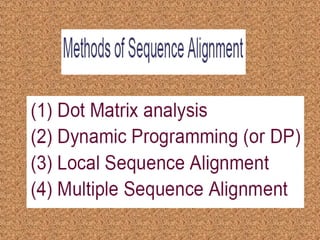
This document outlines a study that will retrieve DNA sequences from GenBank, align the sequences using tools like ClustalW, and identify software that can convert the alignments to Microsoft Word format. It will compare the accuracy of different sequence alignment tools and construct a phylogenetic tree. It then describes CEDIT and SNUFER software. CEDIT uses Microsoft Word's color formatting to identify conserved DNA sequences across an alignment. SNUFER automatically generates tables of single nucleotide polymorphisms from a ClustalW alignment and can export the alignment or table to Word, Excel, or text files. The document demonstrates using SNUFER to load sequences, view an alignment, export it to Word, and view the aligned sequences in Word.