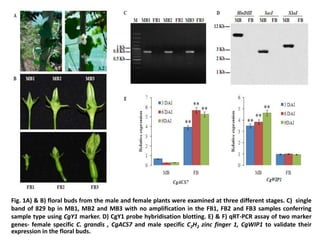
1) The document analyzes microRNA expression in the dioecious plant Coccinia grandis to understand sex differentiation. Small RNA sequencing of male and female floral buds identified 142 microRNAs, including 41 differentially expressed between sexes.
2) Target prediction and expression analysis of microRNAs and their targets revealed roles for certain microRNAs in flower development and sexual dimorphism in C. grandis, including 8 conserved and 8 novel microRNAs linked to sex determination.
3) Reciprocal expression patterns of 16 microRNAs and their predicted targets across development of male and female buds provided evidence of microRNA regulation of genes important for sex differentiation in this species.