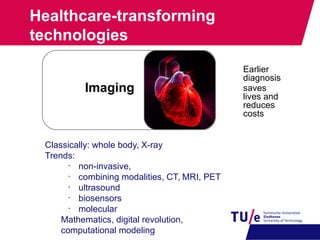
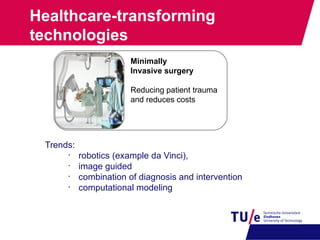
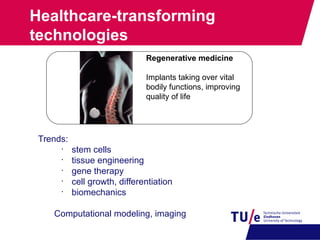
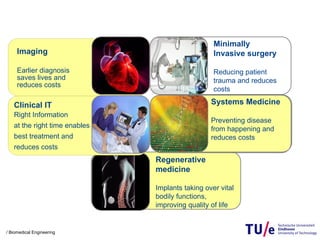
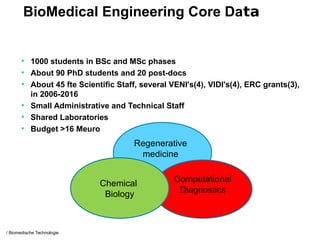
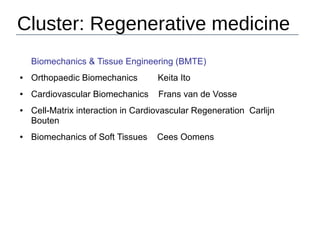
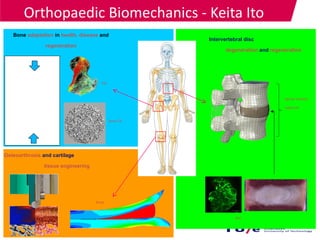
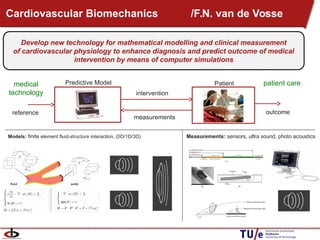
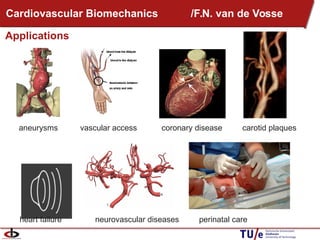
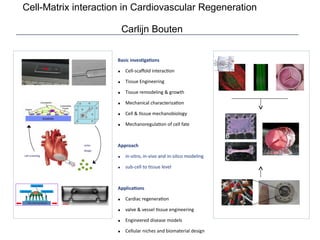
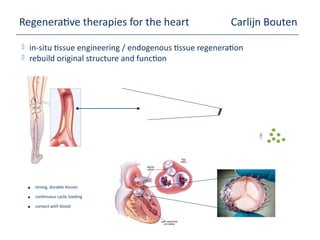
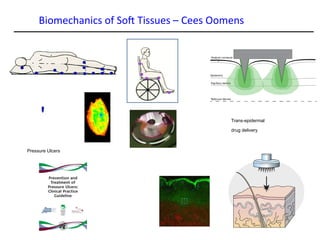
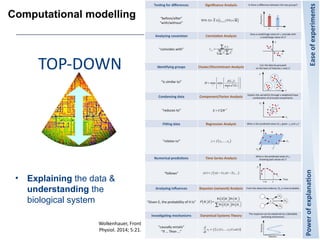
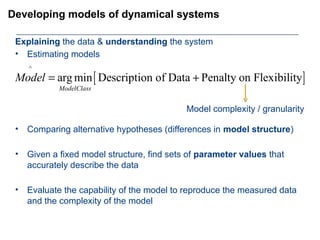
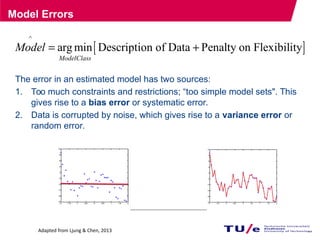
This document provides an overview of the Department of Biomedical Engineering at Eindhoven University of Technology. It discusses general trends in healthcare technology and computational biology examples. The department focuses on areas like regenerative medicine, chemical biology, computational diagnostics, and biomechanics & tissue engineering. Research groups within the department work on topics such as cardiovascular biomechanics, cell-matrix interaction, molecular biosensing, and medical image analysis. The document also provides information on the department's educational programs, collaborations, budgets, and key personnel.




















![Disease progression and treatment of T2DM
• 1 year follow-up of treatment-naïve T2DM patients (n=2408)
• 3 treatment arms: monotherapy with different hypoglycemic agents
– Pioglitazone – insulin sensitizer
• enhances peripheral glucose uptake
• reduces hepatic glucose production
– Metformin - insulin sensitizer
• decreases hepatic glucose production
– Gliclazide - insulin secretogogue
• stimulates insulin secretion by the pancreatic beta-cells
56
FP
G
[m
m
ol/
L]
Schernthaner et al, Clin. Endocrinol. Metab. 89:6068–6076 (2004)
Charbonnel et al, Diabetic Med. 22:399–405 (2004)](https://image.slidesharecdn.com/openlecturepeterhilbers-160719130602/85/Open-Lecture-Peter-Hilbers-July-8-2016-56-320.jpg)