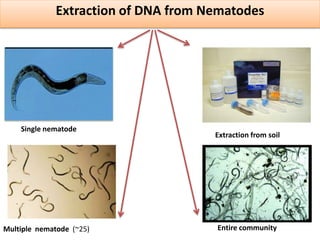
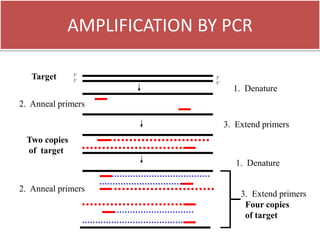
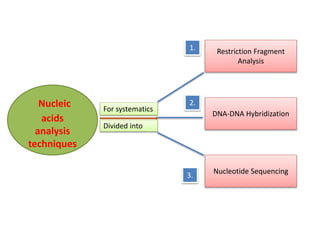
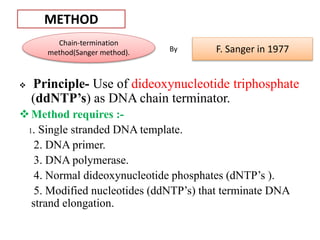
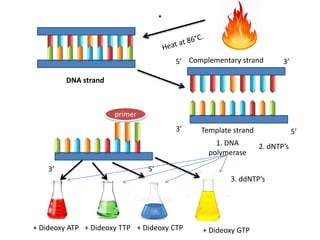
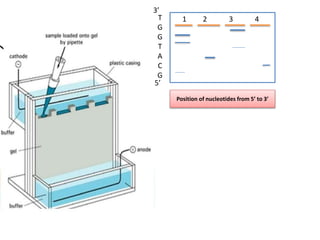
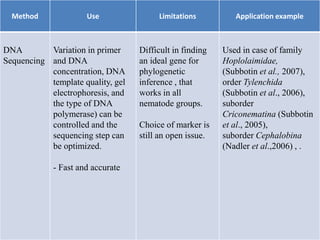
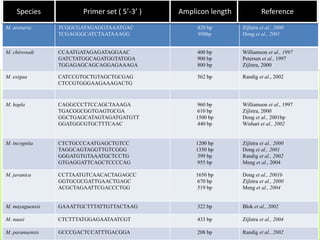
1. The document discusses various molecular techniques used in nematode systematics, including nucleic acid extraction, amplification via PCR, restriction fragment analysis, DNA-DNA hybridization, and nucleotide sequencing.
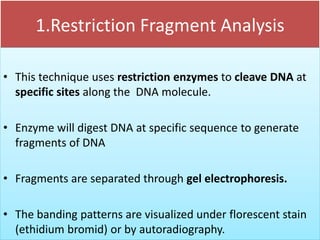
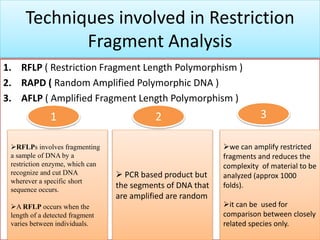
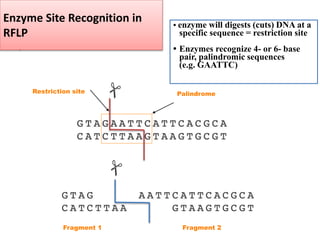
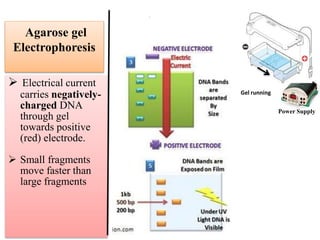
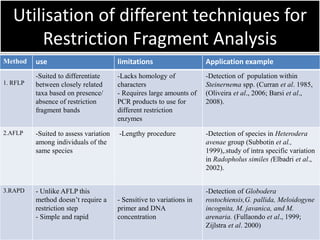
2. Restriction fragment analysis techniques like RFLP, AFLP, and RAPD involve fragmenting DNA with restriction enzymes and comparing fragment patterns to determine relationships between taxa.
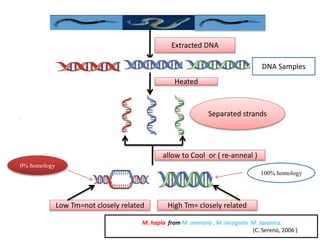
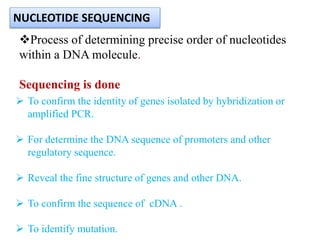
3. DNA-DNA hybridization and nucleotide sequencing provide more precise data for constructing phylogenetic trees and understanding evolutionary relationships between nematode species.