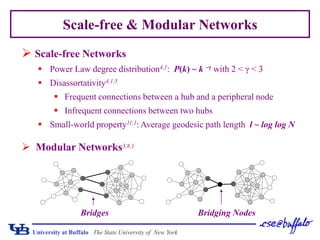
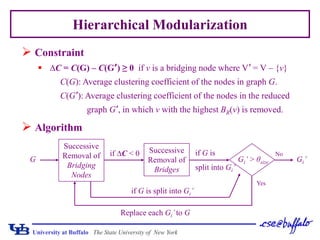
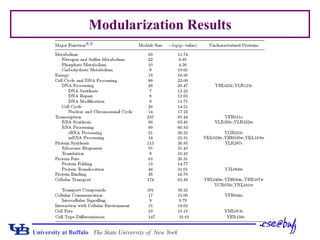
This document presents a method for assessing hierarchical modularity in protein interaction networks. The method involves successively removing bridging nodes and bridges from the network to split it into hierarchical modular subgraphs. Applying the method to a protein interaction network of Saccharomyces cerevisiae revealed four zones of node removal corresponding to bridging, interconnecting, core, and peripheral nodes. Analysis found that bridging and interconnecting nodes were less lethal but more important than peripheral nodes, while core nodes were the most lethal. The modularization results divided the network into hierarchical communities.