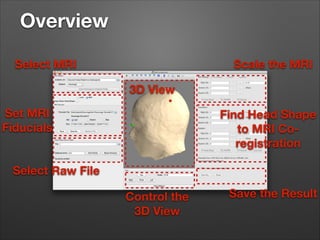
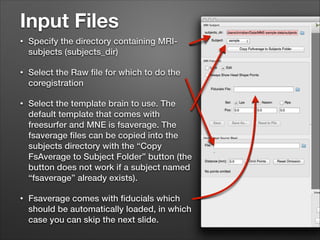
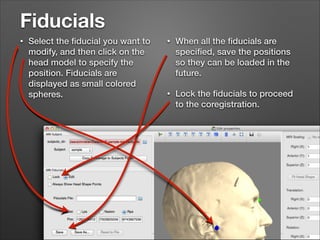
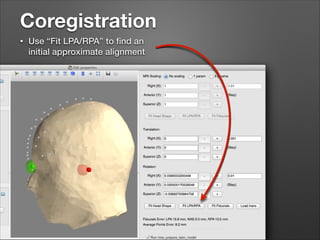
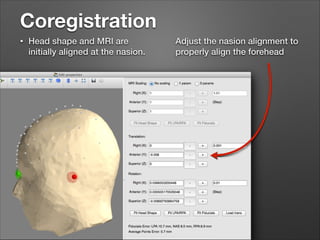
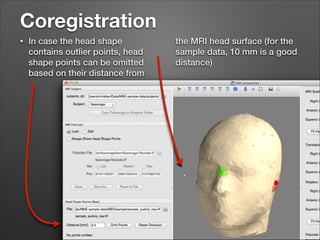
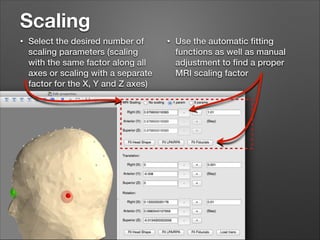
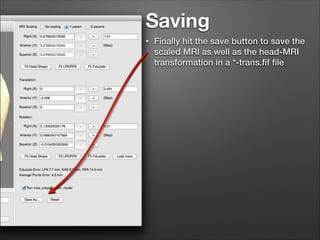
This document provides instructions for coregistering MRI data to MEG data in MNE-Python without a subject's individual MRI. It describes selecting a template brain MRI, setting fiducials, initially aligning the head shape to the MRI, scaling the MRI for proper fit, fine-tuning the alignment manually, and saving the transformed MRI and head-MRI coregistration parameters.