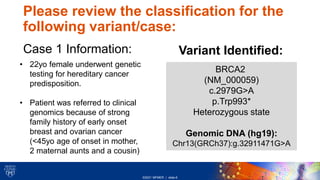
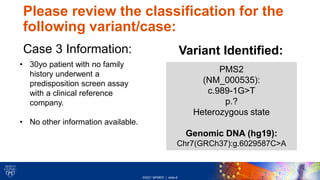
This document provides an overview of a clinical variant interpretation lab exercise. It discusses the objectives of reviewing automated and manual variant classification challenges and examples. The lab will use the American College of Medical Genetics (ACMG) guidelines framework for variant interpretation. Students will review each variant classification criteria discussed in class and verify the preliminary classification given by the Franklin system. Three variant cases are presented for review, involving genes BRCA2, LDLR, and PMS2 in patients with histories of cancer or no reported issues.