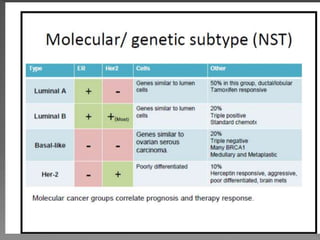
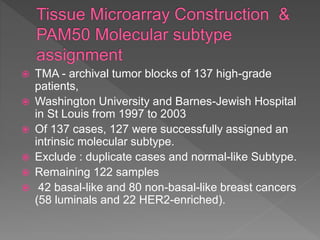
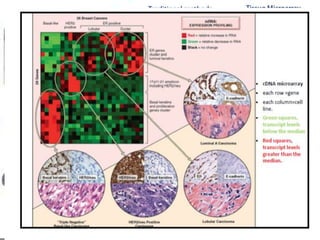
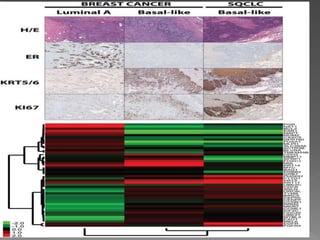
1. The study aimed to validate immunohistochemical (IHC) surrogates for intrinsic breast cancer subtypes identified by gene expression profiling, using the PAM50 assay as the gold standard.
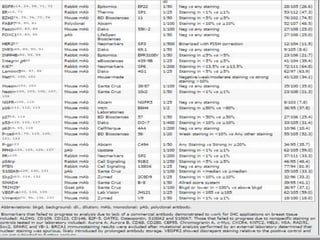
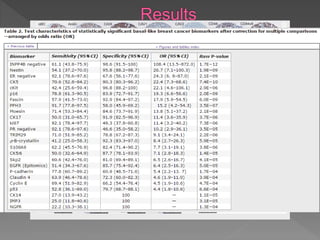
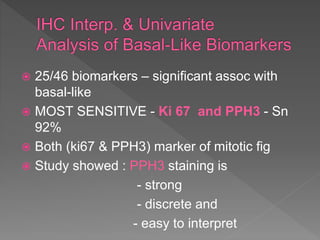
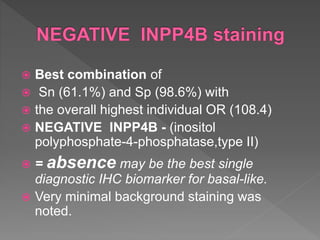
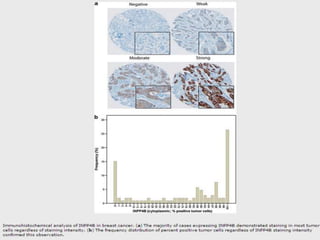
2. Among 46 biomarkers tested by IHC on a tissue microarray, loss of INPP4B expression showed the best combination of sensitivity (61.1%) and specificity (98.6%) for basal-like breast cancer defined by PAM50.
3. Nestin positivity also strongly correlated with basal-like subtype, consistent with other studies, and may be a useful positive basal marker. A multi-marker IHC panel may better define basal-like breast cancer than single biomarkers.