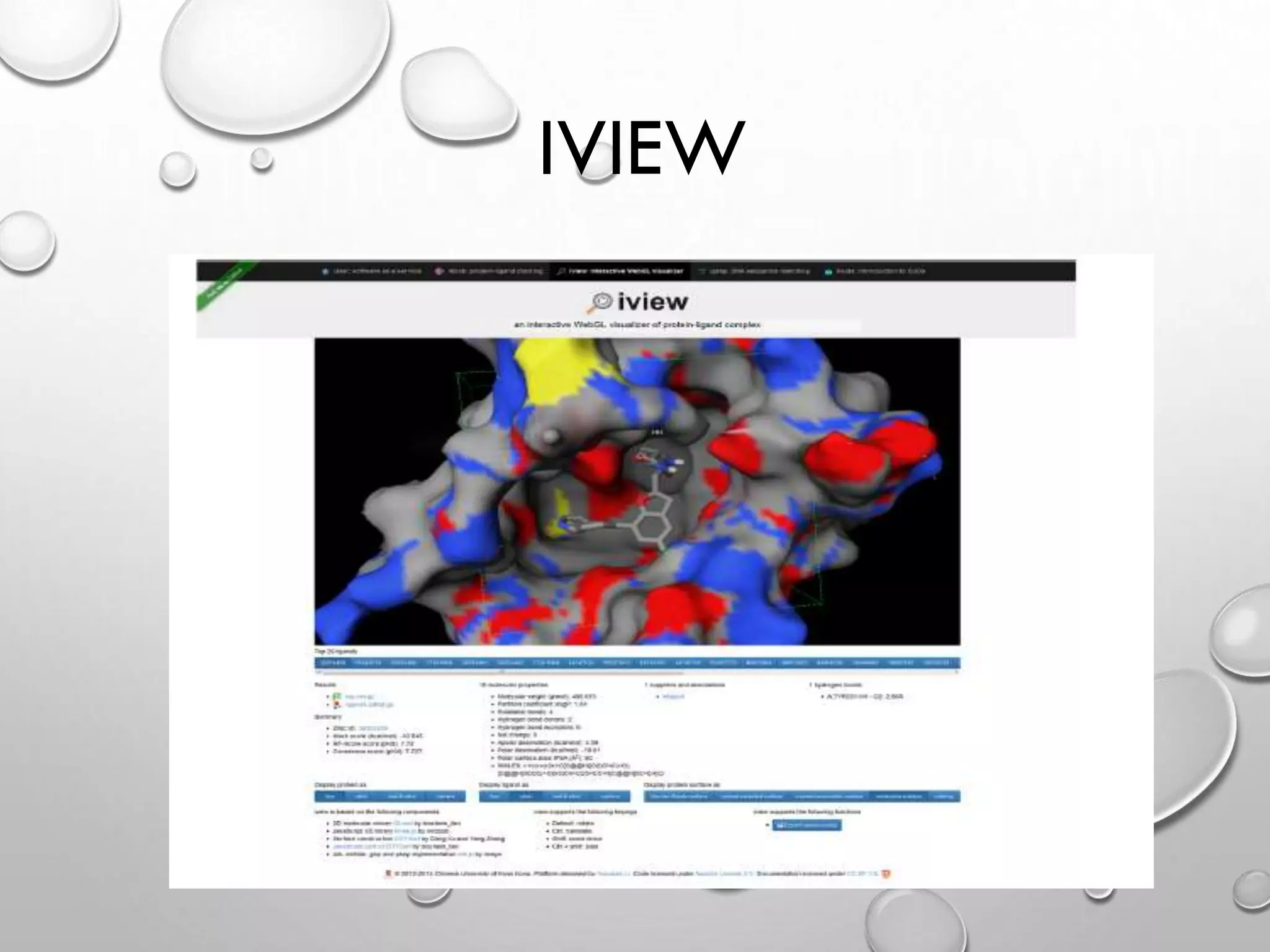
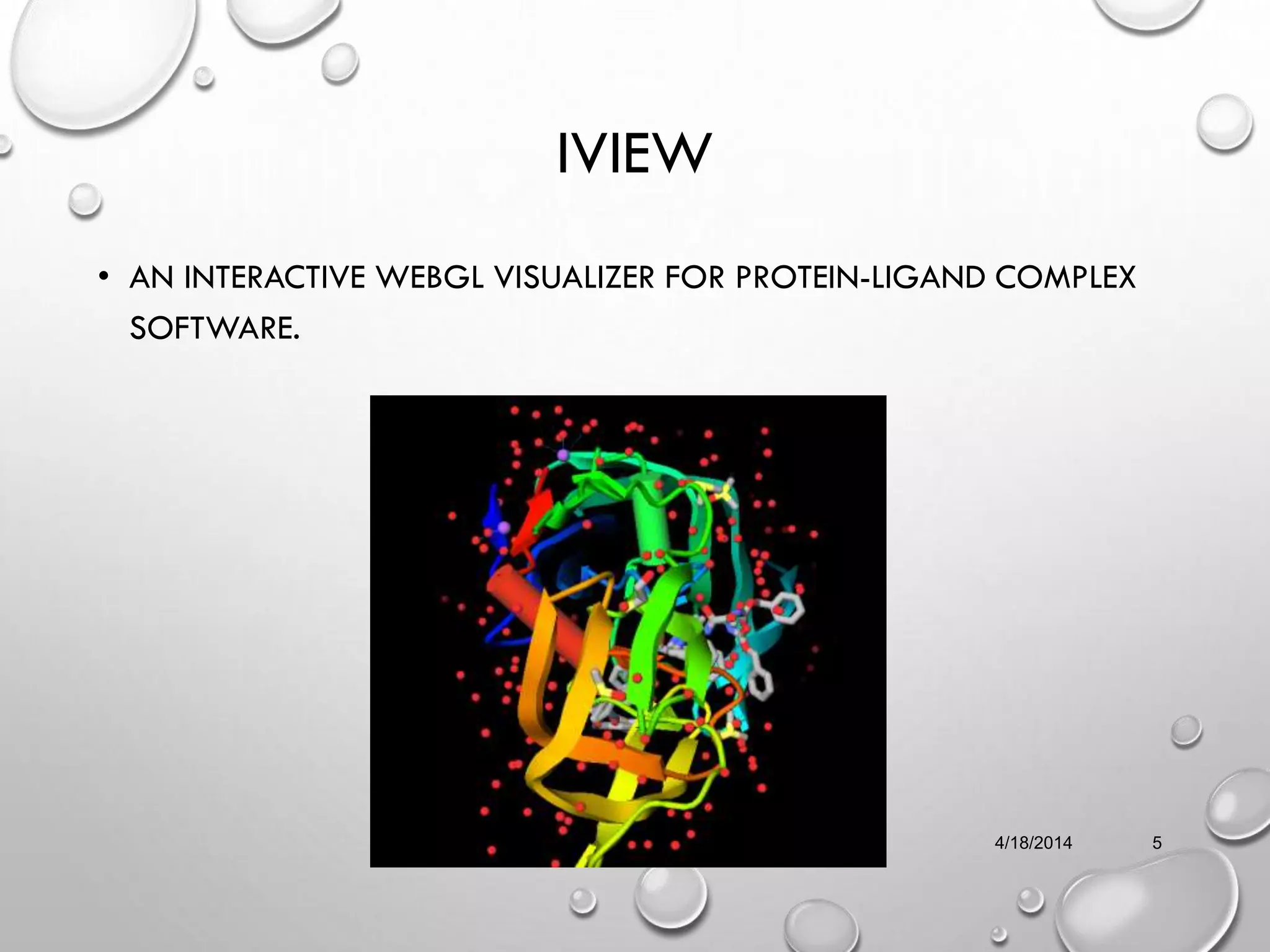
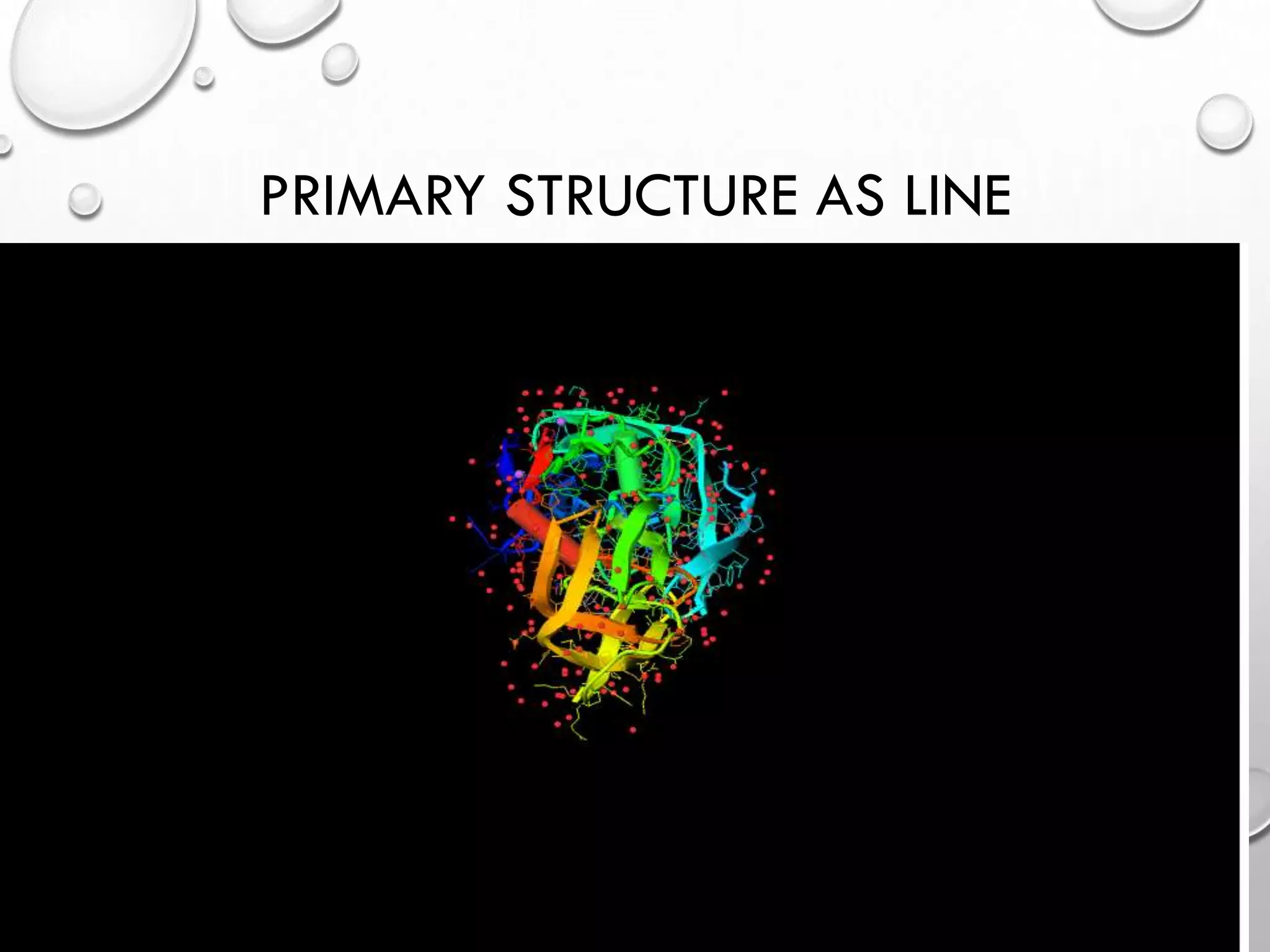
The document describes IVIEW, an interactive WebGL visualizer for protein-ligand complexes. IVIEW allows users to visualize protein-ligand structures and interactions in 3D in their web browser. It features tools like multiple visualization modes, camera controls, labeling, and virtual reality effects. IVIEW aims to provide an easy and accessible way for non-experts to understand protein-ligand binding without requiring specialized software. The tool reads common file formats and is compatible with various browsers and devices.











































![REFERENCES
• HUMPHREY W, DALKE A, SCHULTEN K: VMD: VISUALMOLECULAR DYNAMICS.
• JMOL GRAPH 1996, 14(1):33–38.
• 2. PETTERSEN EF, GODDARD TD, HUANG CC, COUCH GS, GREENBLATT D. M,
• MENG EC, FERRIN TE: UCSF CHIMERA - A VISUALIZATION SYSTEM FOR
• EXPLORATORY RESEARCH AND ANALYSIS. J COMPUT CHEM 2004,
• 25(13):1605–1612.
• 3. MORRIS GM, HUEY R, LINDSTROM W, SANNER MF, BELEW RK, GOODSELL
DS,
• OLSON AJ: AUTODOCK4 AND AUTODOCKTOOLS4: AUTOMATED DOCKING
WITH
• SELECTIVE RECEPTOR FLEXIBILITY. J COMPUT CHEM 2009, 30(16):2785–2791.
• 4. TROTT O, OLSON AJ: AUTODOCK VINA: IMPROVING THE SPEED AND
ACCURACY
• OF DOCKING WITH A NEW SCORING FUNCTION, EFFICIENT OPTIMIZATION,
AND
• MULTITHREADING. J COMPUT CHEM 2010, 31(2):455–461.
• 5. LI H, LEUNG K-S, WONG M-H: IDOCK: AMULTITHREADED VIRTUAL
SCREENING
• TOOL FOR FLEXIBLE LIGAND DOCKING. IN 2012 IEEE SYMPOSIUM ON
• COMPUTATIONAL INTELLIGENCE IN BIOINFORMATICS AND COMPUTATIONAL
BIOLOGY
• (CIBCB). SAN DIEGO, CALIFORNIA, USA: IEEE; 2012:77–84.
[HTTP://IEEEXPLORE.
• IEEE.ORG/XPL/ARTICLEDETAILS.JSP?ARNUMBER=6217214]
• 6. TSE C-M, LI H, LEUNG K-S, LEE K-H, WONG M-H: INTERACTIVE DRUG
DESIGN IN
• VIRTUAL REALITY. IN 15TH INTERNATIONAL CONFERENCE ON INFORMATION
• VISUALISATION (IV). LONDON, UK: IEEE; 2011:226–231.
[HTTP://IEEEXPLORE.IEEE.
• ORG/XPL/ARTICLEDETAILS.JSP?ARNUMBER=6004005]
• 7. STIERAND K, RAREY M: POSEVIEW -MOLECULAR INTERACTION PATTERNS
AT A
• GLANCE. J CHEMINFORM 2010, 2:50.
• 8. LASKOWSKI RA, SWINDELLS MB: LIGPLOT+:MULTIPLE LIGAND-PROTEIN
• INTERACTION DIAGRAMS FOR DRUG DISCOVERY. J CHEM INFORMMODELING
2011,
• 51(10):2778–2786.
• 9. KASAHARA K, KINOSHITA K: GIANT: PATTERN ANALYSIS OF MOLECULAR
• INTERACTIONS IN 3D STRUCTURES OF PROTEIN-SMALL LIGAND COMPLEXES.
• BMC BIOINFORMATICS 2014, 15(1):12.
• 10. HANSON RM, PRILUSKY J, RENJIAN Z, NAKANE T, SUSSMAN JL: JSMOL
AND THE
• NEXT-GENERATION WEB-BASED REPRESENTATION OF 3DMOLECULAR
• STRUCTURE AS APPLIED TO PROTEOPEDIA. ISR J CHEM 2013, 53(3–4):207–
216.
4/18/2014 58](https://image.slidesharecdn.com/iview-140418094140-phpapp02/75/Iview-bioinformatics-tool-CADD-DOCKING-58-2048.jpg)

