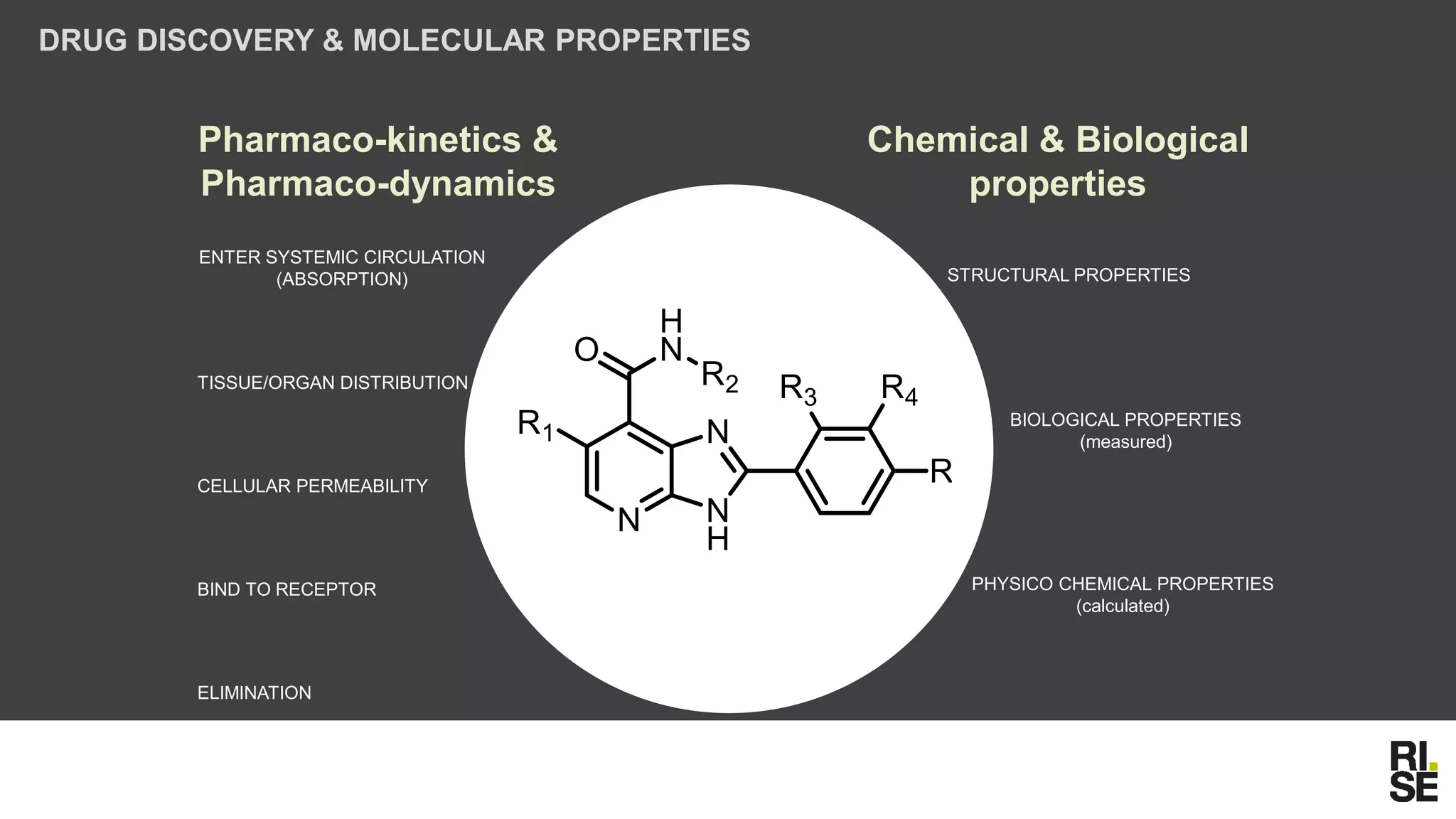
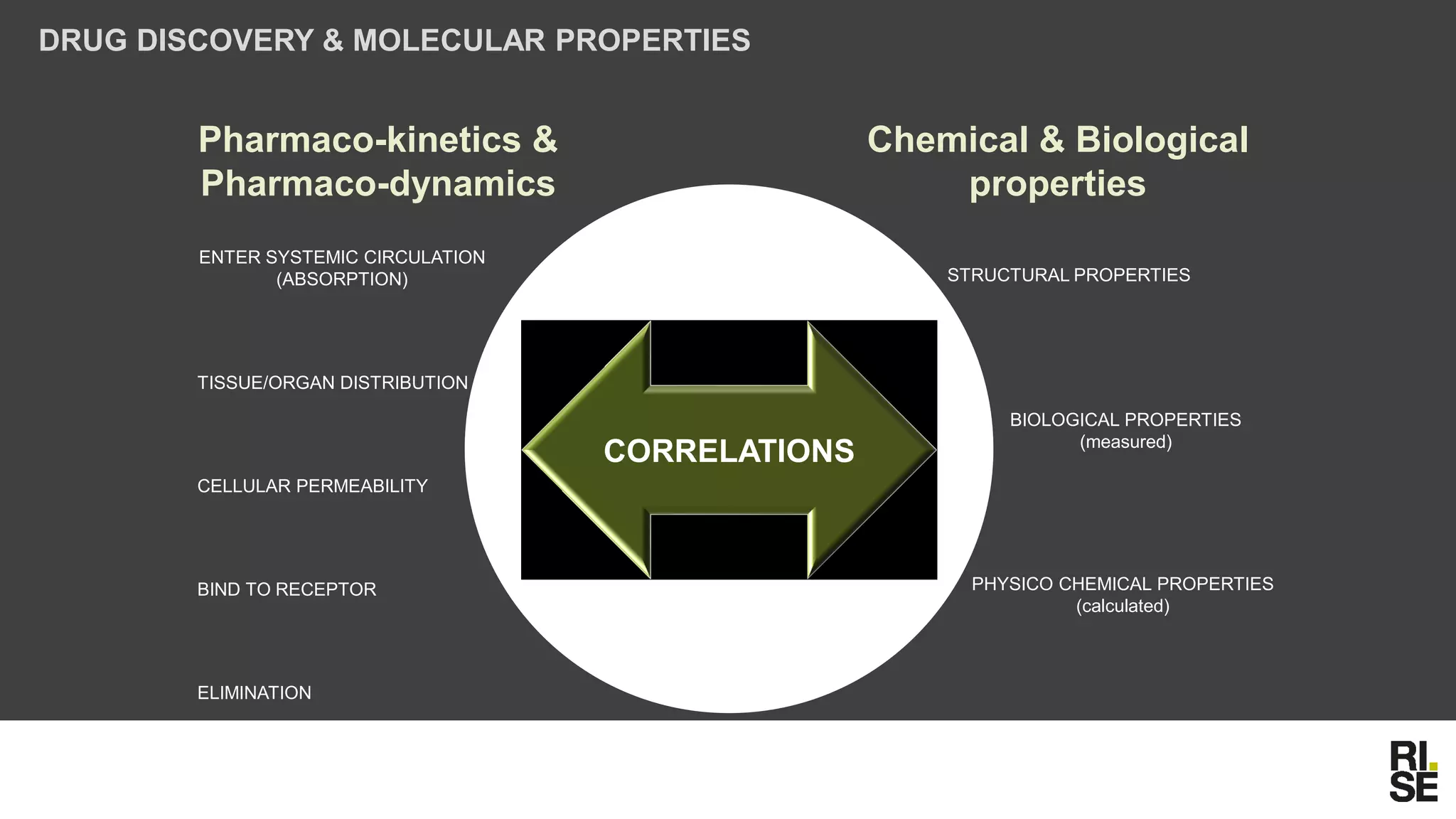
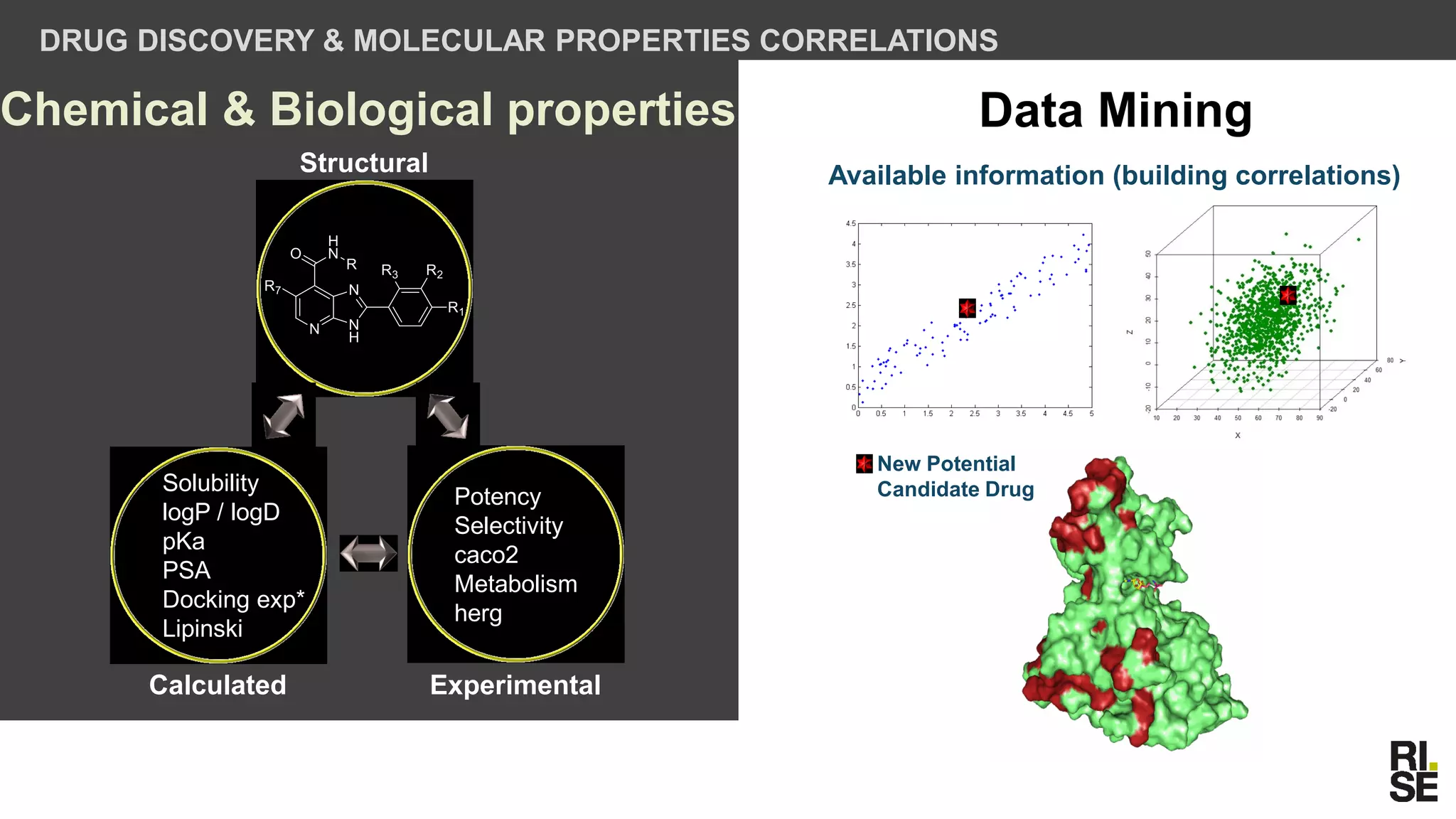
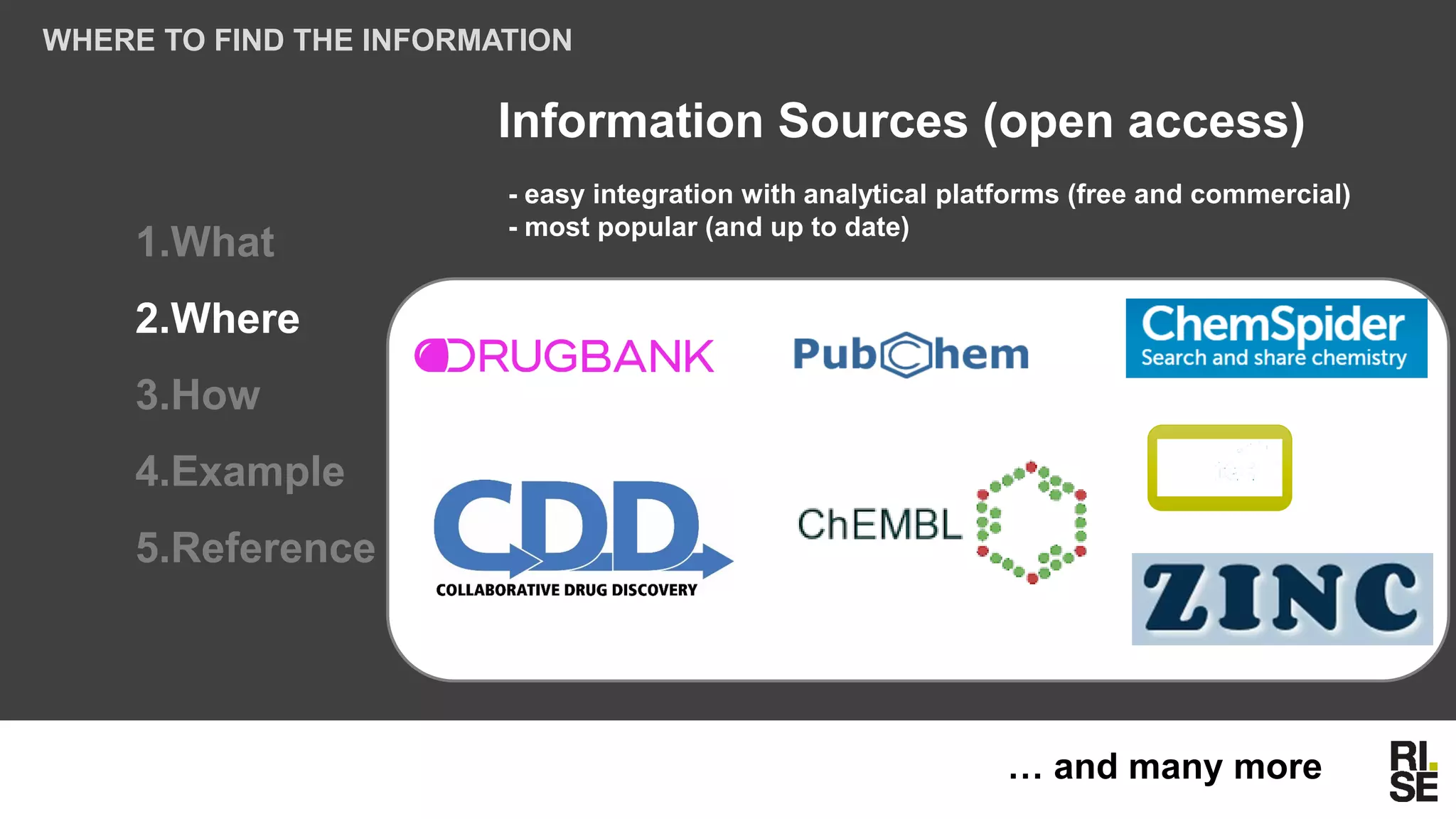
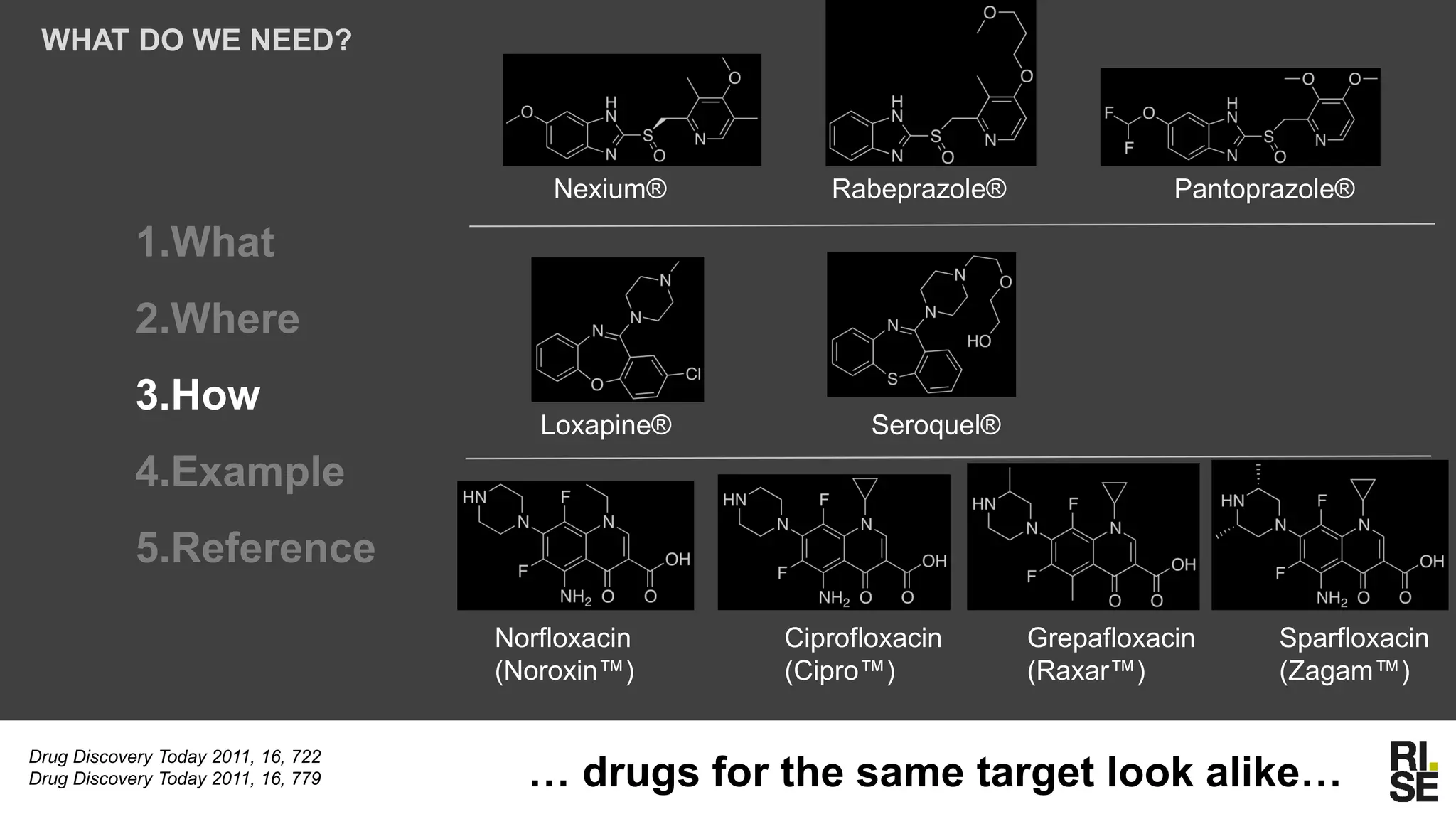
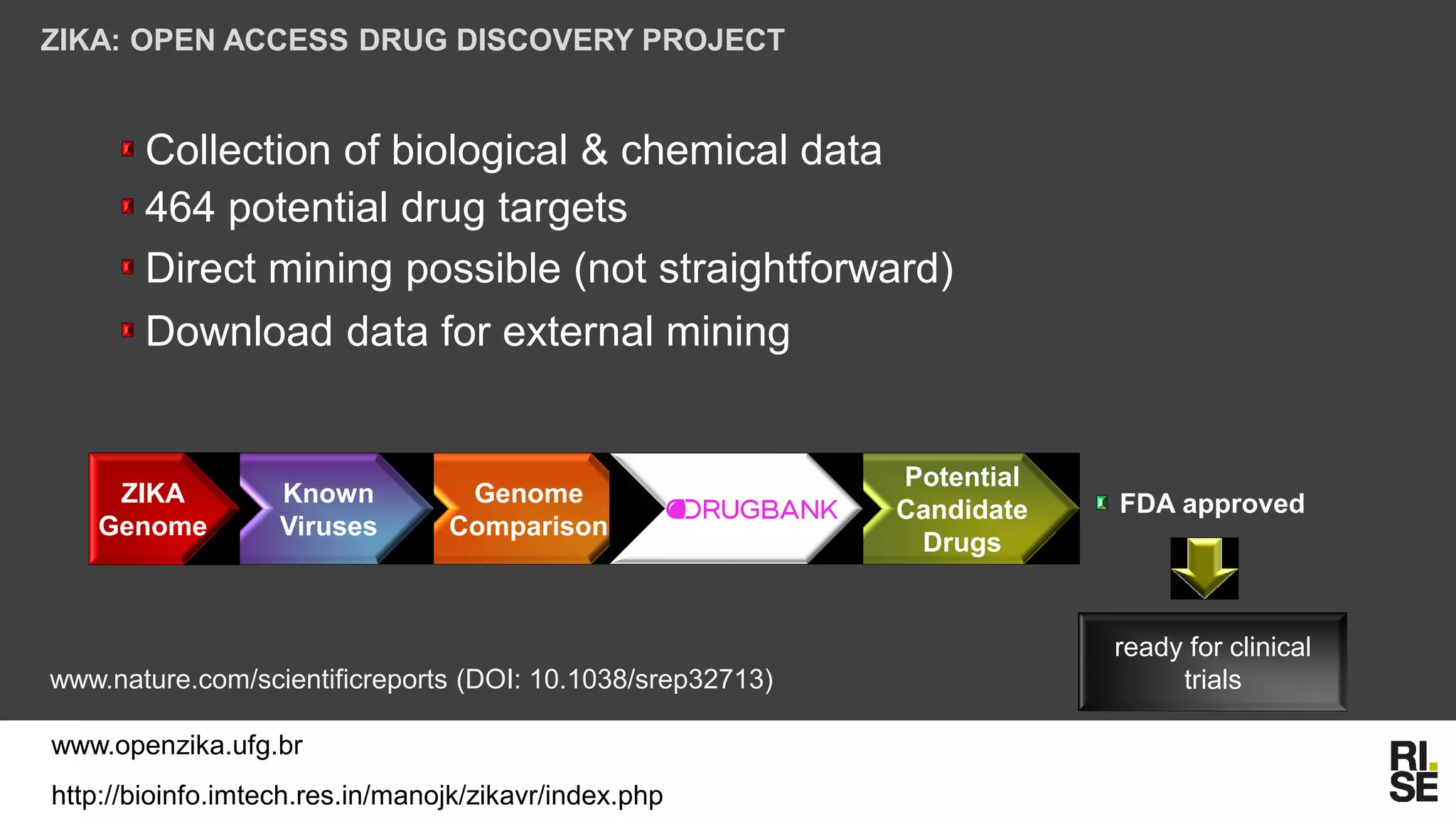
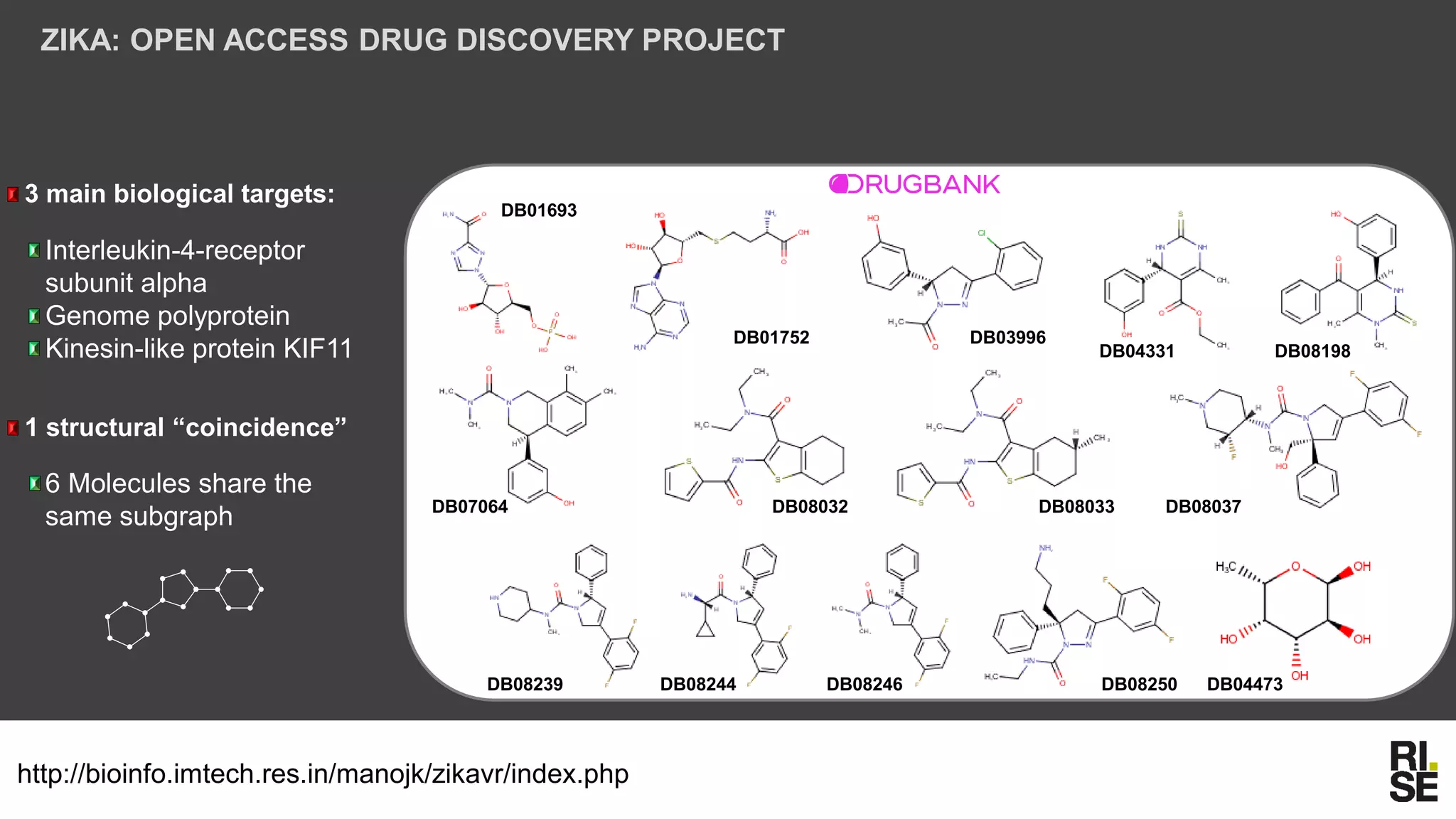
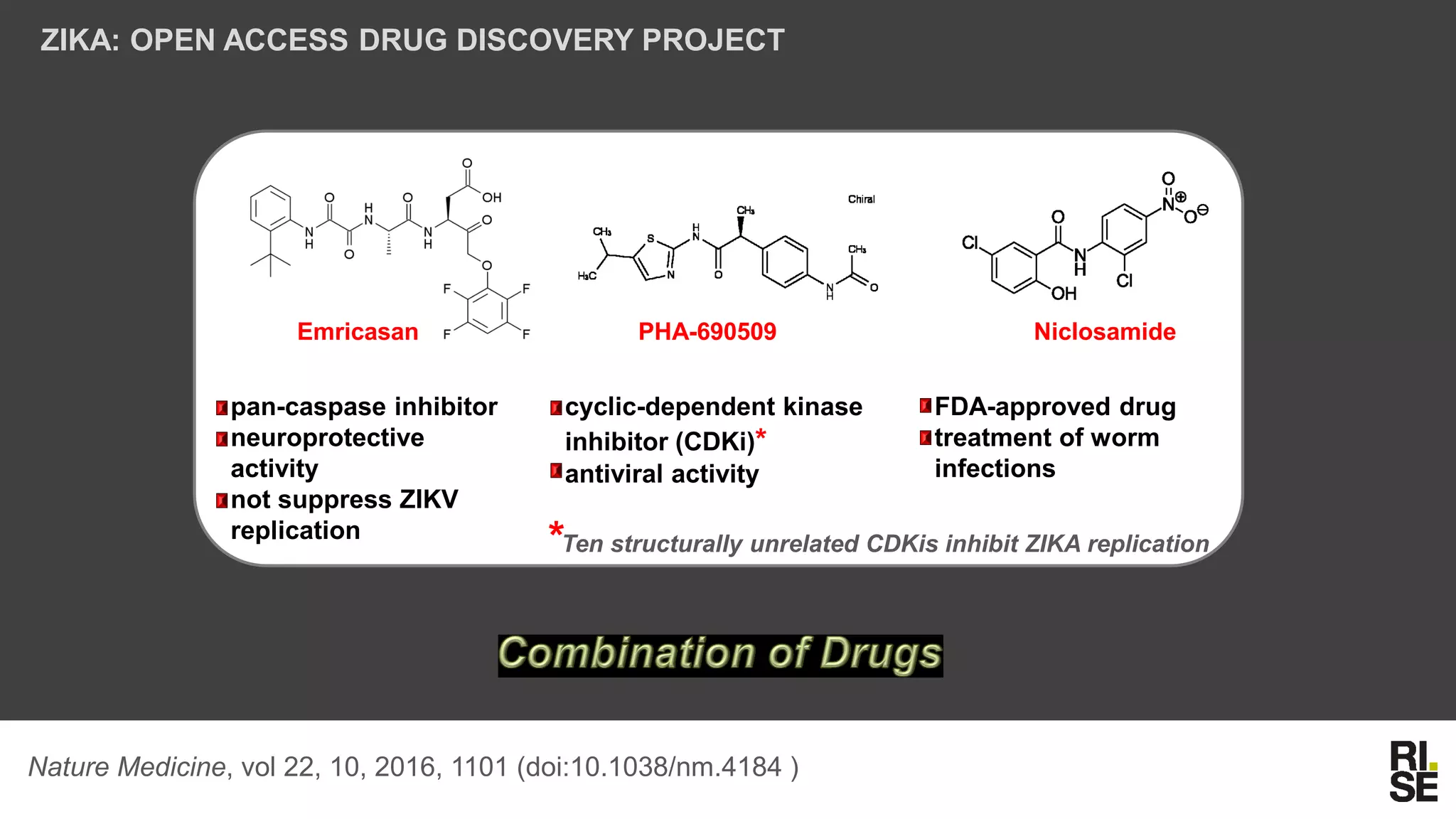
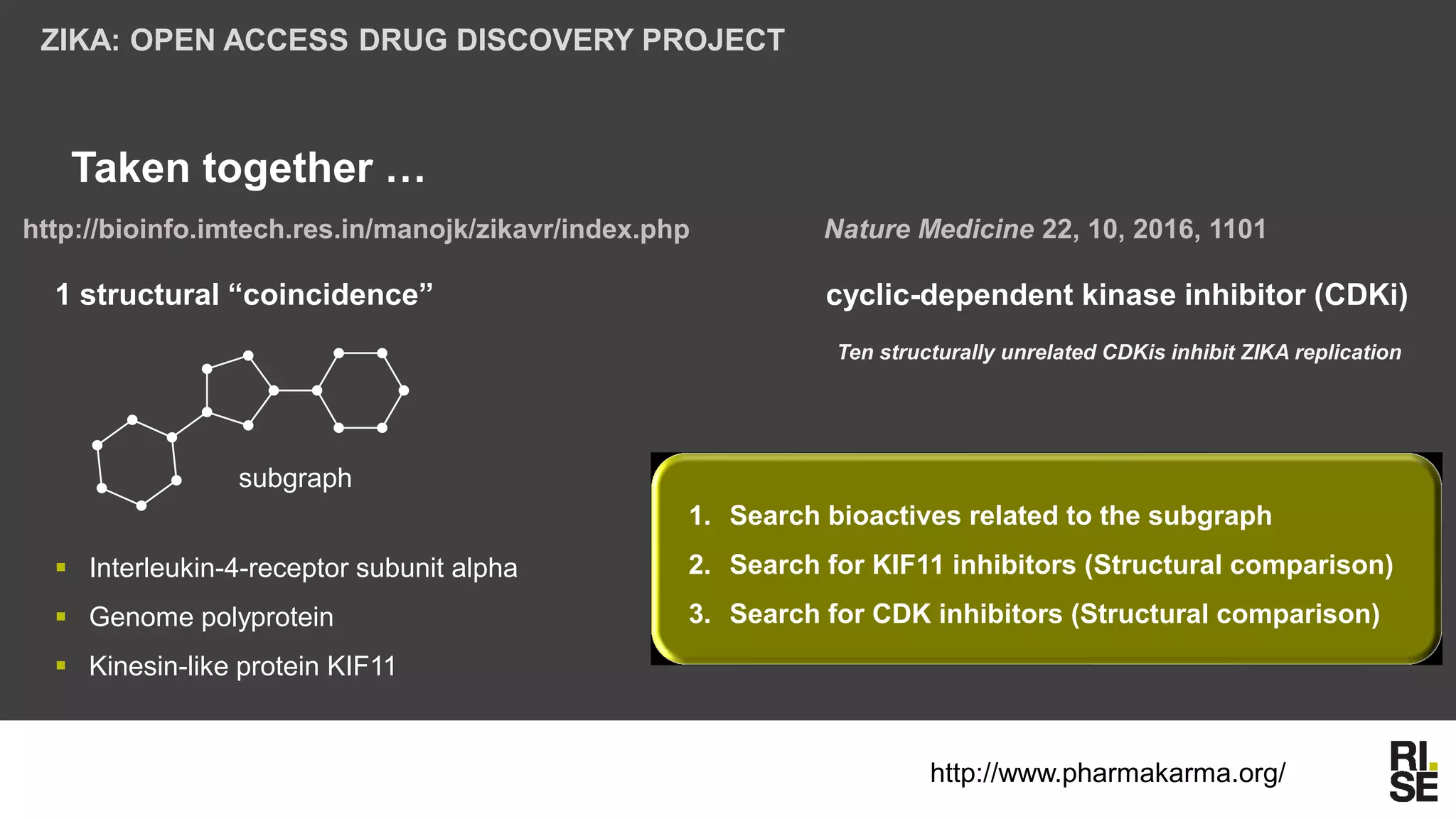
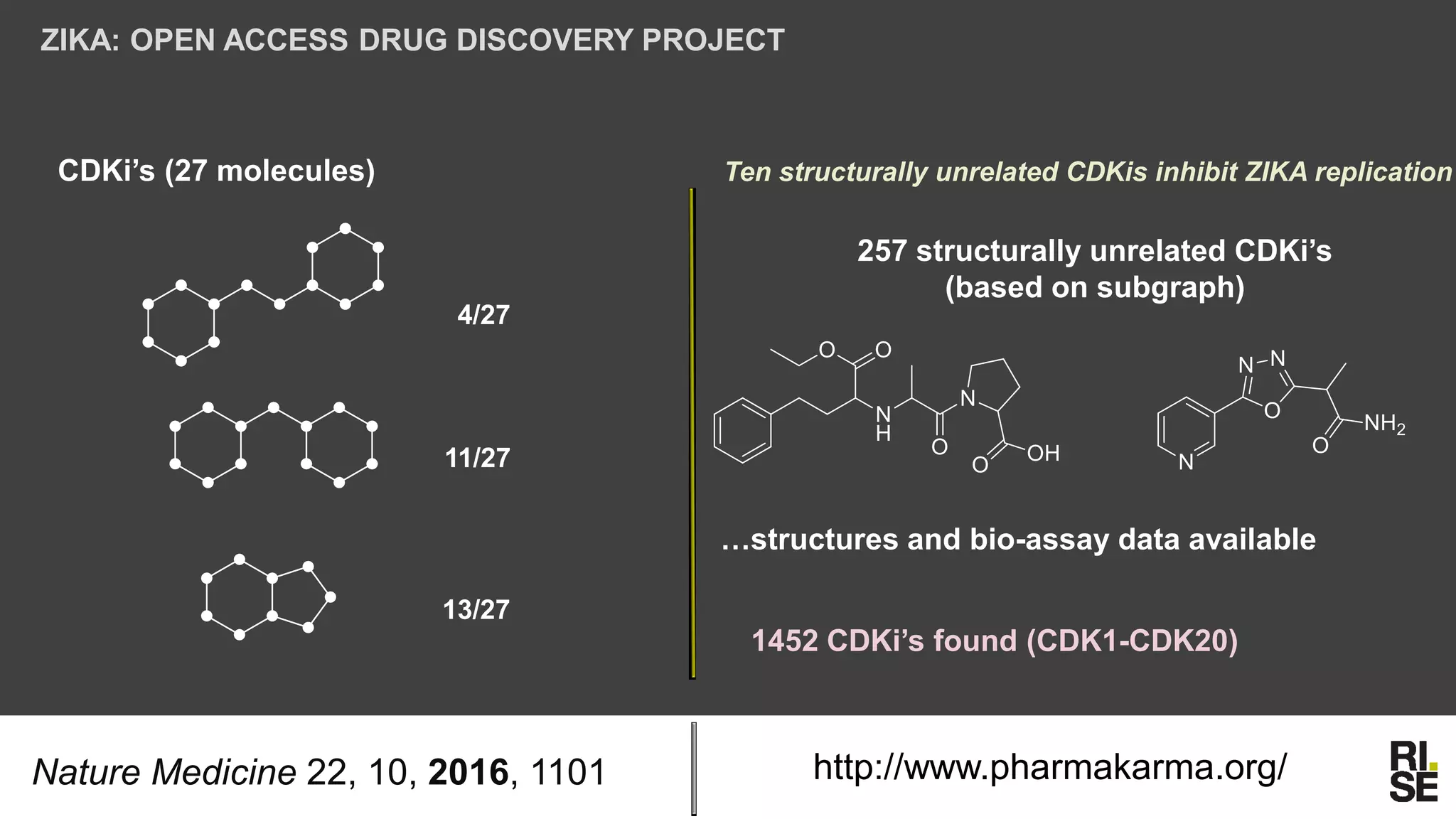
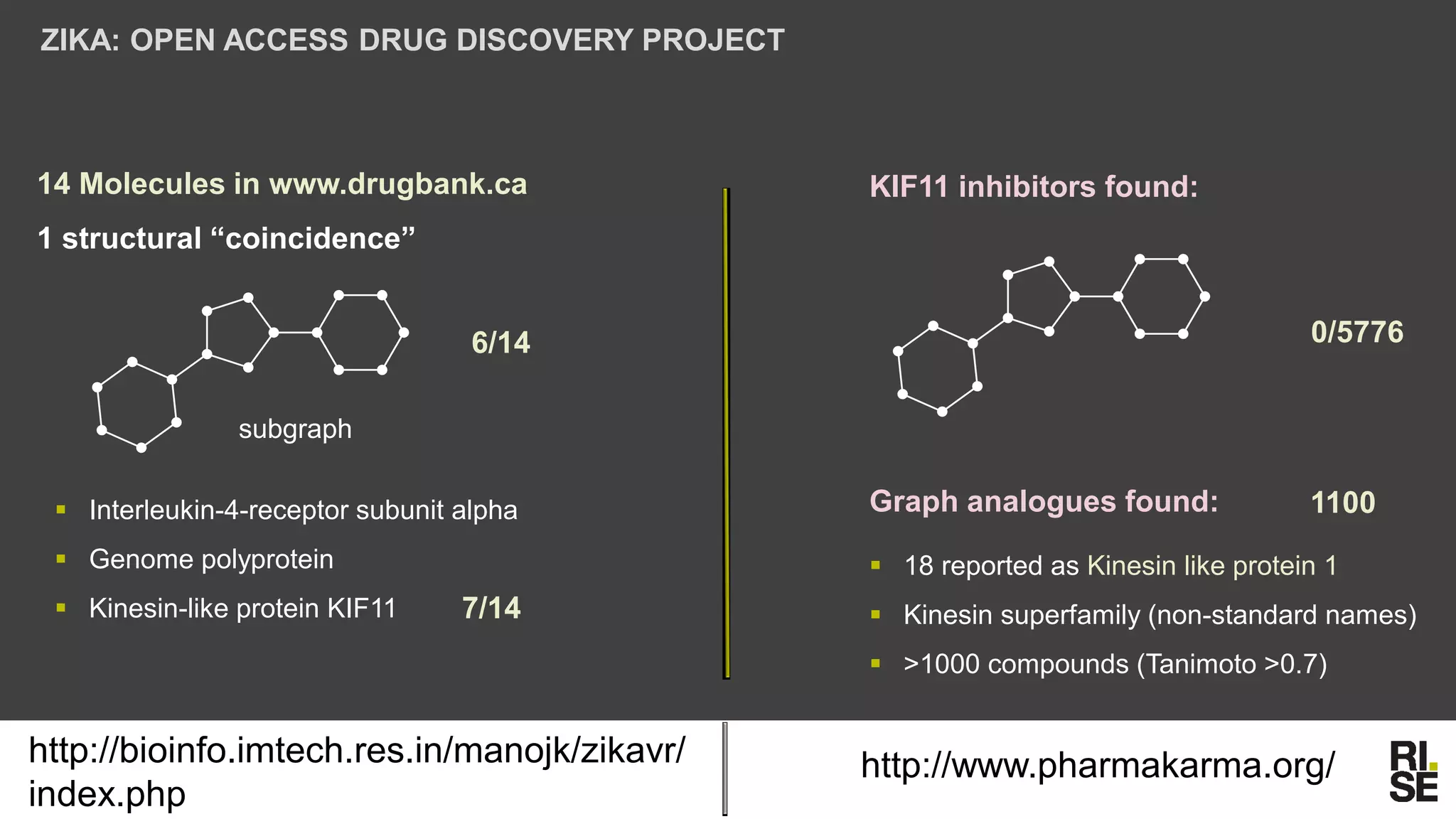
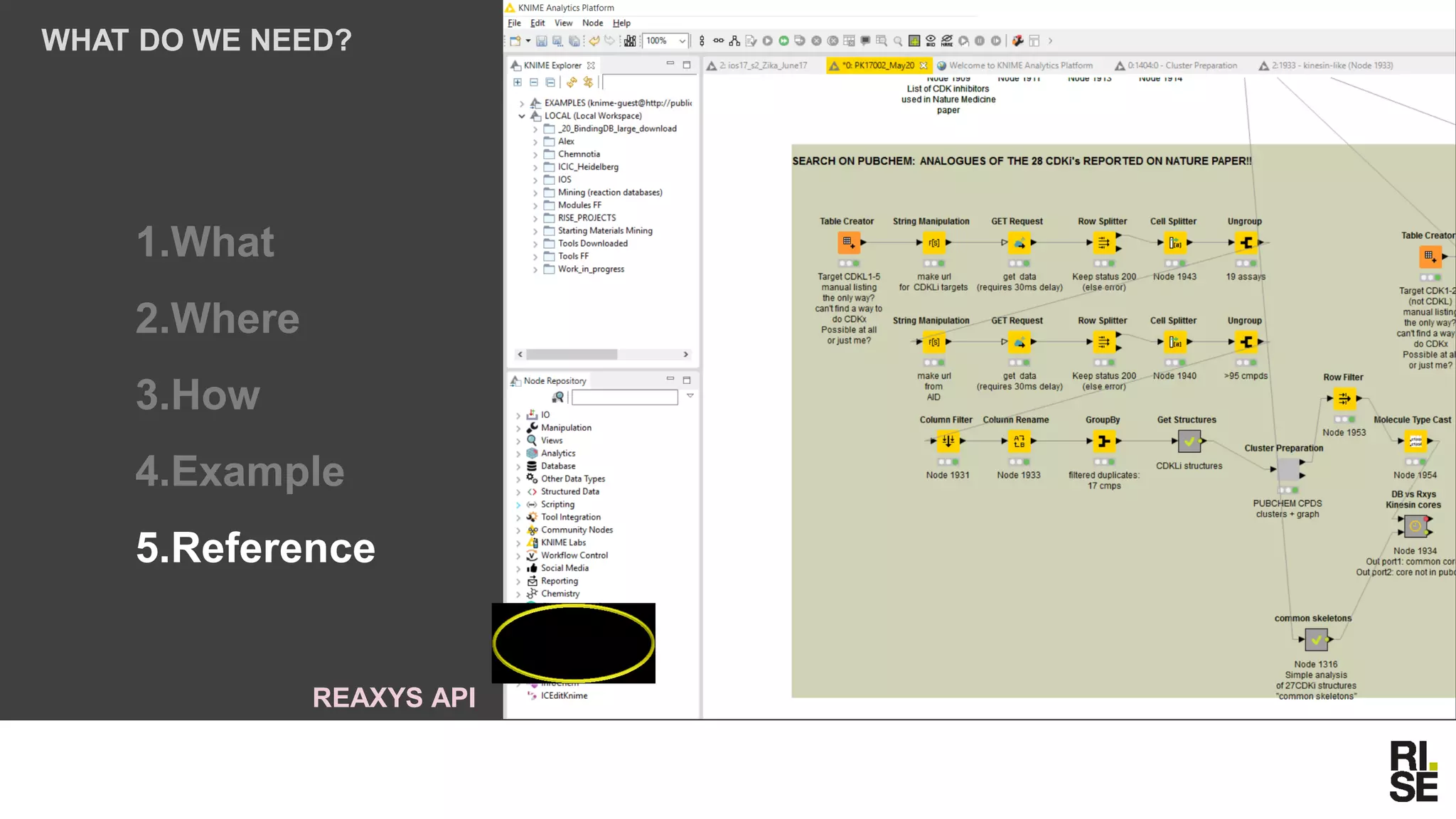
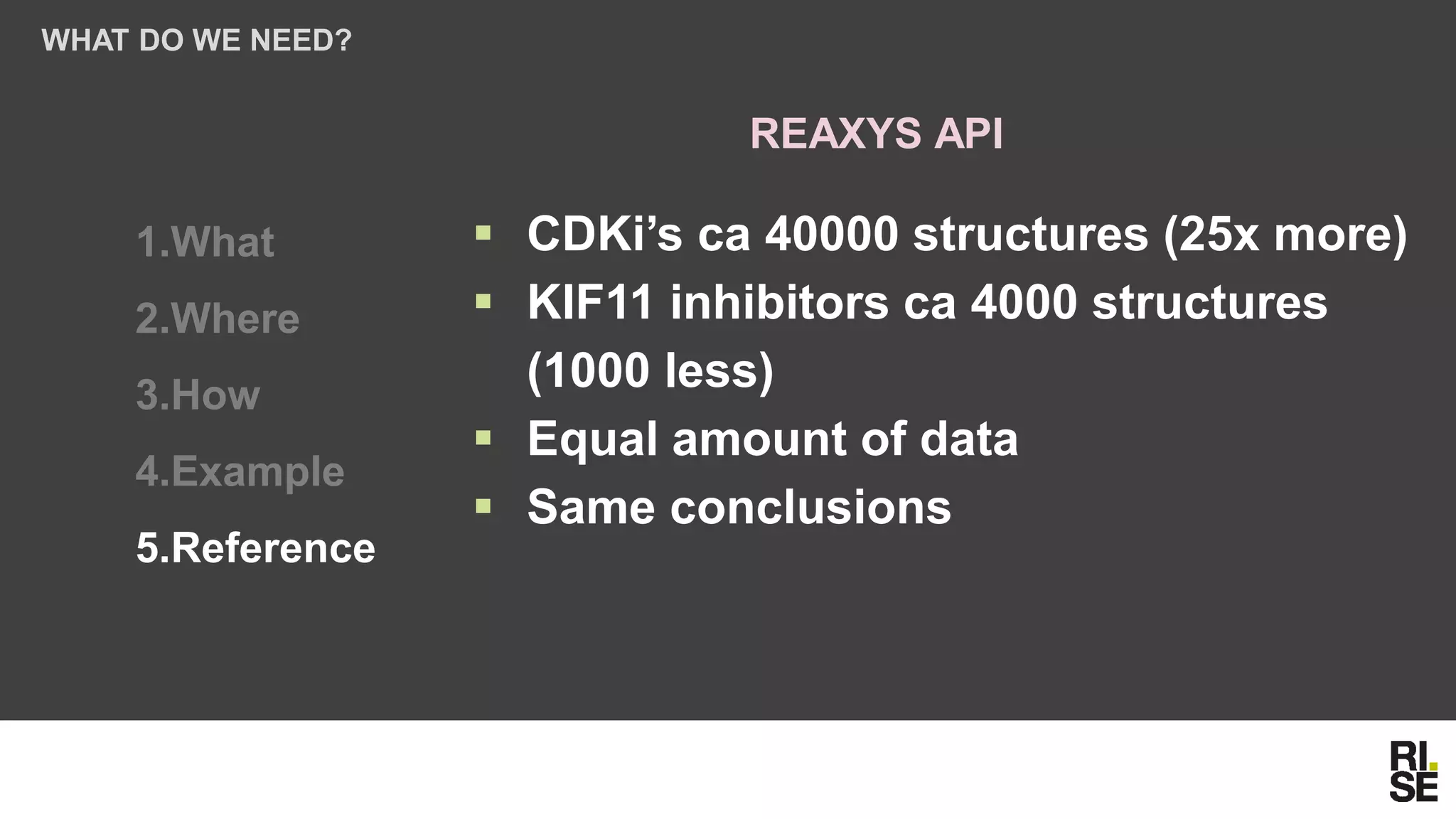
The document discusses the potential of using free and open-access databases in drug design and discovery, highlighting various information sources and the integration of analytical platforms. It emphasizes the importance of data mining and visualization in identifying candidate drugs, particularly in the context of the Zika virus. Ultimately, it suggests that while challenges exist, utilizing accessible information for drug design projects is feasible.