Embed presentation
Download to read offline



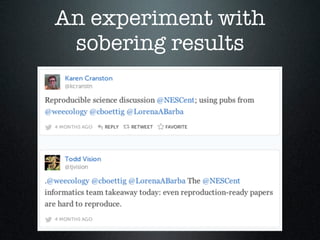








The document discusses a reproducible science panel from 2014 that focused on developing curriculum and addressing challenges in reproducible research within the biological sciences. Key issues include difficulties in software dependencies, documentation effectiveness, and variability in reproducibility practices among researchers. It emphasizes the importance of making research reproducible for facilitating future work and collaboration in scientific communities.











