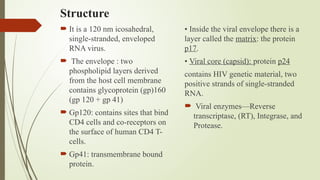
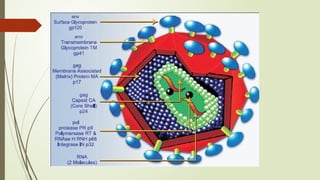
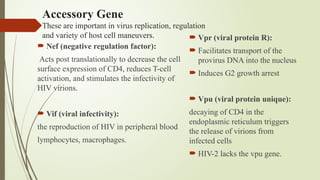
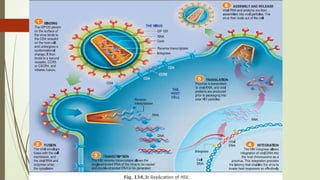
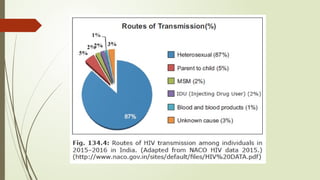
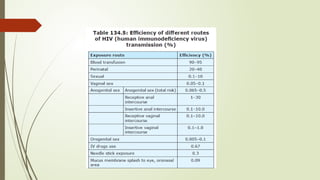
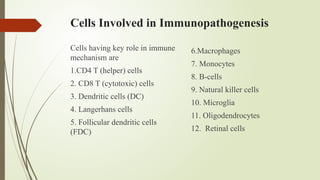
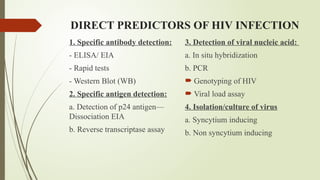
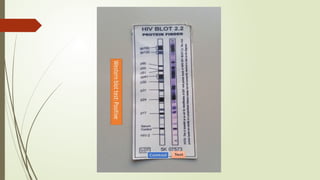
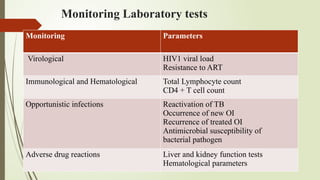
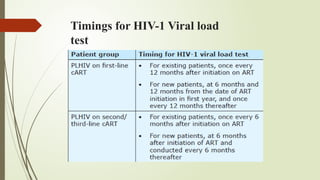
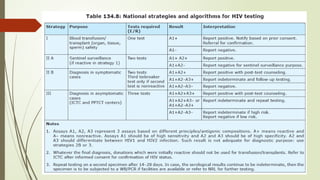
The document provides an overview of human immunodeficiency virus (HIV) virology, including its structure, genetic composition, replication cycle, modes of transmission, and diagnostic testing methodologies. It highlights the distinctions between HIV-1 and HIV-2, their respective disease progression characteristics, and the importance of accurate diagnosis for treatment. Furthermore, it covers laboratory testing techniques, the significance of monitoring parameters, and strategies for enhancing HIV testing services.